Seeking Answers from Big Data in the Era of Precision Medicine
Cancer data can be fragmented and compartmentalized, and many stakeholders are trying to overcome the challenges this poses for advancing research forward. To accelerate progress, cancer researchers need access to curated data from across many different institutions. Establishing an infrastructure to help researchers store, analyze, integrate, access, and visualize large amounts of biological data and related information is the focus of bioinformatics.
Bioinformatics uses advanced computing, mathematics, and different technological platforms to physically store, manage, analyze, and understand the data.
Currently, researchers use many different tools and platforms to store and analyze biological data, including data from whole genome sequencing, advanced imaging studies, comprehensive analyses of the proteins in biological samples, and clinical annotations.
It is often difficult to integrate and analyze data from these various platforms, however, and often researchers don't have access to the raw or primary data created by other studies or lack the computational tools and infrastructure necessary to integrate and analyze it.
In recent years, there has been a boom in the use of virtual repositories—or "data clouds"—to integrate and improve access to research data. Many of these efforts are still in their early stages, and questions remain about the optimal way to organize and coordinate clouds and their use.
NCI's Role in Cancer Bioinformatics
NCI has played a leading role in advancing the science of genomics, proteomics, imaging, and metabolomics, among other areas, to increase our understanding of the molecular basis of cancer.
The NCI Center for Biomedical Informatics and Information Technology (CBIIT) oversees the institute’s bioinformatics-related initiatives.
The National Cancer Informatics Program (NCIP) is involved in numerous research areas, including genomic, clinical, and translational studies, and is exploring how to improve data sharing, analysis, and visualization. For instance, NCIP operates NCIP Hub, a centralized resource designed to create a community space to promote learning and the sharing of data and bioinformatics tools by cancer researchers. NCIP Hub itself is an experiment to see if the cancer research community finds the social and community aspects of the program useful for team science and multi-investigator research teams.
The Cancer Data Science Laboratory (CDSL), in NCI's Center for Cancer Research, develops and uses computational approaches to analyze and integrate laboratory and patient data from cancer genomics and other "omics" research. These computational approaches and algorithms can then be used to address fundamental research questions about the origin, evolution, progression, and treatment of cancer.
Under The Cancer Genome Atlas (TCGA), a research program that was supported by NCI and the National Human Genome Research Institute, researchers have conducted comprehensive molecular analyses of more than 11,000 patients using tumor and healthy tissue samples. More than 1,000 studies have been published based on TCGA-collected data.
Similarly, under NCI's Therapeutically Applicable Research to Generate Effective Treatments (TARGET) program, researchers have identified genetic alterations in pediatric cancers, most of which are from children in clinical trials conducted by the Children's Oncology Group.
NCI’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) is a collaborative consortium of institutions and investigators that perform genomic and proteomic analyses to better understand the molecular basis of cancer. Proteomics data generated by CPTAC research projects are made publicly available in a repository that is accessible by the global research community.
Data from these initiatives and other NCI-supported studies have helped researchers better understand the biology of different cancers and identify potential new targets for therapies.
In some respects, however, these studies have only scratched the surface of what can be learned from the vast amount of data collected as part of this research. As a result, there has been a new push in the research community to find ways to make these data, and the tools to analyze them, more widely accessible.
Democratizing Big Data for Cancer Research
As a federal agency, NCI is uniquely positioned to democratize access to cancer research data. NCI's Office of Data Sharing (ODS) coordinates data sharing policies across NCI and the cancer research community. ODS manages NCI data submissions and access to online databases, provides education and outreach for NCI data sharing policies, and examines the uptake and use of NCI data.
NCI has launched several initiatives to provide researchers with easier access to data from TCGA, TARGET, and other NCI-funded research, and the resources to analyze the data.
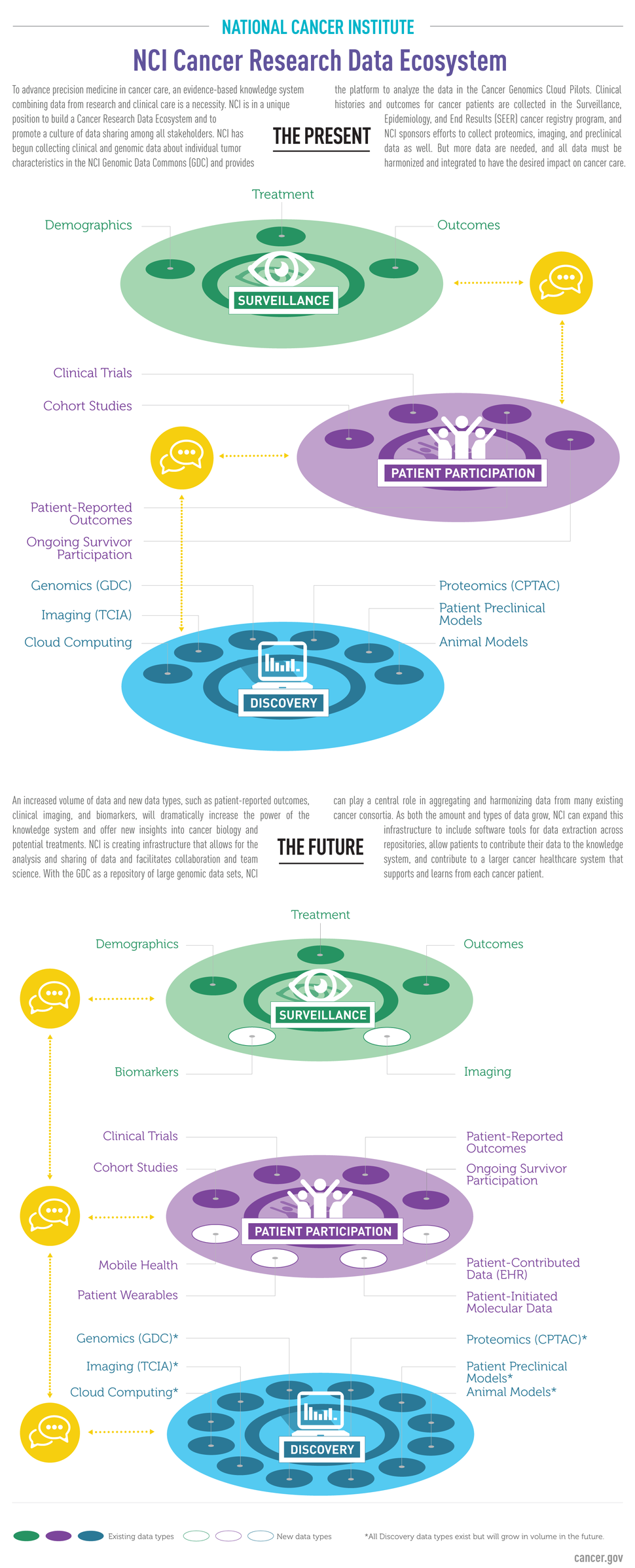
The NCI Cancer Research Data Commons (CRDC) is a data science infrastructure that connects cancer research data collections with analytical tools. The CRDC can be used to store, analyze, share, and visualize cancer research data types, including proteomics, animal models, and epidemiological cohorts. The CRDC includes the NCI Genomic Data Commons, the NCI Cloud Resources, the Data Commons Framework, and other projects.
- The NCI Genomic Data Commons (GDC) provides a single source for data from NCI-funded initiatives and cancer research projects, as well as the analytical tools needed to mine them. The GDC includes data from TCGA, TARGET, and the Genomics Evidence Neoplasia Information Exchange (GENIE). The GDC will continue to grow as NCI and individual researchers and research teams contribute high-quality, harmonized data from their cancer research projects. The GDC provides the cancer genomics repository for projects falling under the NIH Genomic Data Sharing Policy.
- The GDC's Data Analysis, Visualization, and Exploration (DAVE) tools make it easy for cancer researchers to work with GDC data by bringing the information technology infrastructure required for downloading, storing, and analyzing big data directly to researchers. Experts in diverse areas of biology and other disciplines can also use DAVE tools to incorporate cancer genomic data into their research.
- NCI provides resources that use cloud technology to provide researchers with access to genomic and other data from NCI-funded studies. These NCI Cloud Resources are used to explore innovative methods for accessing, sharing, and analyzing molecular data. Each resource, implemented through commercial cloud providers, operates under common standards but have distinct designs and means of sharing data and analytical tools, with the goal of identifying the most effective means for using cloud technology to advance cancer research.
- The Data Commons Framework provides the core components for building and expanding the CRDC, including services for securing, finding, and annotating data, as well as user workspaces for analyzing data and sharing results.
To enable integration of data from CRDC repositories, a Cancer Data Aggregator (CDA) will support search and analysis across distinct data types. The CDA will allow researchers to combine data from diverse scientific domains and perform integrated analyses which can be shared with collaborators.
Protecting Patient Privacy
An important aspect of data sharing is the ability to link data at the patient level across disparate data sources without exposing identifiable information. NCI is evaluating approaches and creating software for generating unique patient identifiers that can be used to link patient data from different sources without sharing identifiable information beyond the organizations authorized to hold such information. The software-generated identifiers will preserve the privacy of cancer patients who share their data with the cancer research community.