Taking RAS Research to Space
, by Albert Chan
Albert Chan earned his PhD with Robert Clubb at UCLA, and did a postdoc with Karen Anderson at Yale. He is currently a Staff Scientist with Dhirendra Simanshu in the Structural Biology group of the NCI RAS Initiative.
On December 5, 2018, I watched the Falcon 9 rocket launch from Cape Canaveral, Florida, on SpaceX CRS-16 mission. A flash of fire, followed by an unrelenting rumbling sound that crescendoes to a deafening roar, shaking the ground and the air. As I watched the rocket ascending and gradually disappearing from my vision, I prayed for the safe docking of the capsule to the International Space Station (ISS), not just because I wanted to make sure the six astronauts on the ISS get their supply of food and water, but also because I had experiments in the capsule! In collaboration with the International Space Station National Laboratory (ISSNL), the NCI RAS Initiative launched a panel of RAS proteins in various conditions to promote crystal growth in microgravity on the International Space Station (ISS). Our goal was to solve protein structures by X-ray crystallography that we could not solve with crystals grown under Earth’s gravity.
HVR – a missing piece in the current RAS protein structures
Getting protein crystals is the first step in revealing protein structures. There are currently over 300 crystal structures of RAS proteins in the Protein Data Bank (PDB), so what are we trying to achieve by solving another structure? Turned out none of the structures in the PDB shows the full structure of the protein. A RAS protein, such as HRAS, KRAS or NRAS, contains a GTPase-domain (G-domain) core and a short (~20 amino acids long) hypervariable region (HVR). The G-domain binds to effector proteins to activate signal transduction pathways that eventually lead to cell growth and proliferation (cancer if this gets unregulated). The HVR directs and anchors the protein to the cell membrane, the prime location for RAS to interact with its signal transduction partners.
Most of the RAS proteins in the PDB were crystallized with just the G-domain without the HVR. In rare cases where the full protein constructs were used, only the G-domain part of the protein was visible. One important exception is the KRAS-PDEδ complex structure we solved in 2016, in which in one crystal form the HVR adopts an alpha-helical structure (PDB: 5TAR), while in the other crystal form the HVR appears unstructured (PDB: 5TB5). However, what we would like to see is the HVR in KRAS alone, since there are published results and our unpublished data obtained using molecular modelling (in collaboration with Lawrence Livermore National Laboratory) suggest that the HVR interacts with the G-domain prior to translocation to the cell membrane. If we can understand how the HVR interacts with the G-domain at the atomic/structural level, we can potentially design cancer drugs that can capture the protein at this infancy state, so that it can no longer travel to the cell membrane to signal cell growth.
We attempted to crystallize the full-length version (1 to 188 amino acids) of several oncogenic mutants of KRAS in our lab. Unfortunately, and perhaps not too surprisingly, we only saw the G-domain and about a third of the HVR, probably because the HVR is too flexible and is only spending a fraction of the time binding to the G-domain. To increase our chance in observing the full HVR, we included a small molecule that acts as a molecular glue which enhances the binding of the HVR to the G-domain. While this protein-small molecule complex did crystallize, the X-ray diffraction data were uninterpretable due to significant mosaicity (a measure of order in crystals, and this translates to how “messy” the data look) so we could not solve the structure. Around this time, we learned of the opportunity to send experiments to the ISS. Since there is evidence that crystals grown in the low Earth orbit appear to be more perfect – they are bigger, more ordered, and diffract better in general – we decided to give it a shot. The history of microgravity crystallography and the physics behind the improvement of crystal quality (spoiler alert: it has to do with diffusion, convection and gravity), is nicely reviewed by Alexander McPherson and Lawrence DeLucas.
Crystallizing RAS proteins on the International Space Station
We coordinated with Dr. Marc Giulianotti and April Spinale from the ISSNL to include our experiments in the launch. Between our proposal getting awarded mission time and the launch day, we had about five months to develop hardware suitable for transportation and crystallization under microgravity. In previous missions, researchers generally used specialized hardware that could only accommodate a limited number of samples. We decided to modify the normal hardware we use in our lab for our mission so that we could achieve higher sample density (since real estate is at a premium; have you ever seen the inside of the ISS?) and increase our chance in getting crystals. Most of the modifications were done on reagent volume and sealing methods to minimize spilling and withstand freeze-thaw (we would be shipping our samples to the ISS frozen, upon thawing on the ISS crystallization would proceed). In the end, we sent a total of 600 samples. The samples had five weeks to crystallize under microgravity, and they returned to the Earth on January 13, 2019.
Retrieving the samples is probably the riskiest part of the experiment. Protein crystals are sensitive to freezing without special cryo-protection. Therefore, after our protein crystallized on the ISS, they were returned to us at room temperature. That means our samples had to withstand the vibration from re-entry, splashdown, and transportation in sea, land and air. A big part of our hardware modification was done to minimize spillage and crystal damage during transportation. Fortunately, and to our pleasant surprise, all of our samples survived the harsh transportation. More importantly, we found many protein crystals when we examined them in our lab!
What we learned from our crystals grown on the International Space Station
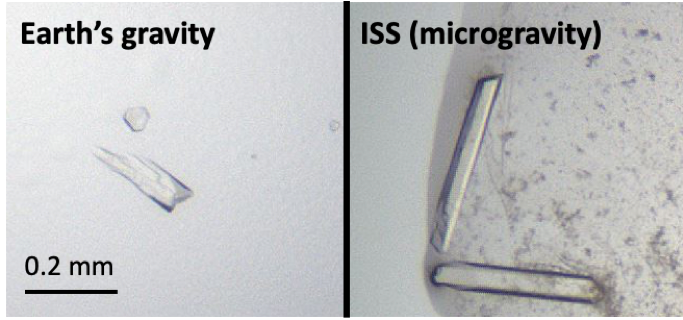
About 25% of the microgravity crystals were visibly bigger and had nicer appearances (smoother surfaces, sharper edges, etc.). The data collected with X-ray was also better. We saw as much as five-fold improvement in signal-to-noise and 50% improvement in mosaicity. This shows that the beauty of our crystals is not only skin-deep; they are beautiful inside as well! With the data quality improvement, we were finally able to solve protein structures that we could not solve with the Earth crystals before, including the small molecule-bound KRAS that suffered from mosaicity problem when crystallized in our lab.
So the experiment was a success… at least partially. While we were excited that we could solve new structures, even in these structures we could not observe the HVR. This could have been mainly due to our crystallization condition (identified in Earth’s gravity), forcing similar crystal packing interactions even in the microgravity environment. Nevertheless, our experiment confirmed what other researchers have seen in microgravity crystals – they are indeed of better quality and might allow structure determination that will not be possible otherwise. If we had another chance to crystallize proteins on the ISS, we would try crystallization screening of RAS proteins or protein complexes using miniaturized nanoliter crystallization trays and for proteins that do crystallize in our lab but suffer mosaicity problems. Given our valuable experience with the ISSNL this past year, I am confident we will be able to unlock more structures in future missions and take RAS research to a new height.