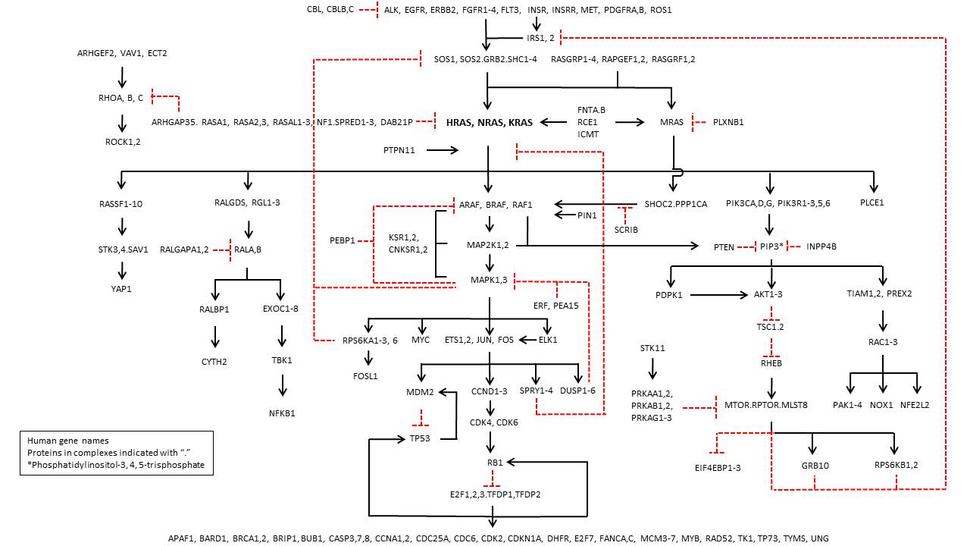
RAS Pathway v2.0
, by Frank McCormick
Many thanks for your valuable input into the RAS pathway diagram. Gaddy Getz and Matt Myerson and colleagues at the Broad Institute along with Bob Stephens, Jim Hartley and colleagues at the Frederick National Laboratory for Cancer Research are utilizing this diagram to develop a RAS-centric version of The Cancer Genome Atlas (TCGA). We hope that this will provide useful data for the RAS community to investigate as well as reveal new and provocative functional relationships.
Our original pathway diagram (posted on October 22, 2014) has been altered incorporating suggestions from the following investigators:
- Phil Stork (stork@ohsu.edu) suggested adding RASGRP1, but but we left out PDZGEF1 which (I think) is a RAP1 GEF, not a RAS GEF. Any input on this would be welcome.
- Julie Irving (julie.irving@ncl.ac.uk) suggested adding FLT3 (also suggested by Kevin Shannon shannonk@peds.ucsf.edu), PTPN11 (also suggested by Ben Braun, UC San Francisco) and CBL. On Ben’s advice, PTPN11 was kept vague with a positive arrow to RAS activation of RAF, without a clear target.
- Manuela Baccarini (manuela.baccarini@univie.ac.at) noted that MAP2K1 activates PTEN by sending it to the plasma membrane. RAF1 inhibits ROCK2, but was not included in our updated diagram because the drawing was getting too complicated. This may be fixed in a subsequent version.
- Mike Nickerson (nickersonml@mail.nih.gov) included negative feedback to IRS1 and 2 from the PI3 kinase pathway.
- Anne Goriely (anne.goriely@imm.ox.ac.uk) suggested adding the MRAS (RRAS3) –SHOC2.PPP1CA pathways that activate RAF kinase and ERF, an ETS-family protein that inhibits ERK1 (aka MAPK3) and 2 (MAPK1).
- Eric Collisson (eric.collison@ucsf.edu) suggested adding the RHO GEF ARHGEF2, aka GEF-H1. Rob Rottopel has recently shown this also acts on the MAPK pathway via interaction with KSR1, but I couldn’t find a way of representing that in this version.
- Jim DeCaprio (james_decaprio@dfci.harvard.edu) suggested adding cyclin D2 (CCND2) and 3 (CCND3) in addition to Cyclin D1 (CCND1) as MAPK targets.
Ping Lu (klu@bidmc.harvard.edu) suggested adding PIN1 as an activator of RAF1 kinase (aka CRAF). It could have been added at many other sites (see Cell Res 24, 1033, 2014 for a review by Zhimin Lu and Tony Hunter). - Michael Tainsky (mat@wayne.edu) noted that AP2 is essential for RAS transformation, but I couldn't find a direct link to the RAS pathway. Any suggestions are welcome.
- Maria Zajac-Kaye (mzajackaye@ufl.edu) suggested adding E2F target genes, including thymidilate synthase (TYMS). Target genes that appear on other parts of the pathway were not listed, to keep it simple.
- Ramon Parsons (ramon.parsons@mssm.edu) suggested adding the RAC GEF, PREX2.
- Shyam Biswal (sbiswal@jhu.edu) and Carola Neumann (neumannc@upmc.edu) suggested adding NFE2L2 (aka NRF2), and NOX1.
- Eric O’Neill (eric.oneill@oncology.ox.ac.uk) suggested adding a mini-version of the Hippo pathway (RASSF, MST [STK3 and 4], SAV1, YAP1).
- Joe Ramos (joeramos@hawaii.edu) and Naoto Ueno (nueno@mdanderson.org) suggested adding PEA15.
- Larry Feig (larry.feig@tufts.edu) suggested adding RASGRF1 and 2.
- Howard Crawford (crawford.howard@mayo.edu) proposed adding VAV1, ECT2 and TIAM1. Howard also suggested DOCK10, but I wasn’t sure where to put it. Suggestions welcome!
- Mike White (michael.white@utsouthwestern.edu) suggested adding EXOC1-8 connected to the RALA, B node and CNKSR1, CNKSR2, SHOC2 connected to the RAF node.
- Karen Cichowski (kcichowski@rics.bwh.harvard.edu) suggested adding DAB21P and INPP4B.
- Andrew Sharrocks (a.d.sharrocks@manchester.ac.uk) suggested adding ELK1.
- Mark Philips (philim01@nyu.edu) suggested adding FNTA and B, RCE1 and ICMT1.
- Debbie Morrison (morrisod@mail.nih.gov) suggested adding a feedback arrow from MAPK1,3 to the RAFs.
- Channing Der (cjder@med.unc.edu) suggested adding MYC as a MAPK1/3 substrate. He also included RGL, RGL2 and RGL3 with RALGDS as another effector that links RAS with RAL. Additionally, the RALGAPs, RALGAPA1 and RALGAPA2, and PLCE1 are included as RAS effectors.
- Philippe Roux (philippe.roux@umontreal.ca) pointed out errors in the pathway as originally drawn: “The pathway should indicate that RPS6KB1 and RPS6KB2 are targets of mTOR (they are the p70 S6Ks). The related p90 S6Ks (RPS6KA1, RPS6KA2, RPS6KA3, RPS6KA6) are actually targets of MAPK1/3, and should be transferred to that branch of the pathway." Steen Hansen (steen.hansen@childrens.harvard.edu) also pointed out these errors and suggested we add FOSL1 downstream of RPS6KA1,2,3 and 6. Additionally, there are two more isoforms of EIF4EBPs, and thus the pathway should indicate EIF4EBP1-3.
- Julian Downward (julian.downward@cancer.org.uk) also noted some errors, “You should have PIK3CA, PIK3CD and PIK3CG here, but not PIK3CB as p110beta does not interact with RAS. Also, for regulatory subunits, there should be PIK3R 1, 2, 3, 5, and 6, but not 4 – this is VPS15, the regulatory partner of VPS34. PIK3R 5 and 6 are the regulatory subunits of p110gamma, so are fine to have here.”
Thank you for these suggestions.
Gene Names, Cognate Proteins, and Aliases in the RAS Pathway
The table comprises the name of each human gene in the RAS Pathway diagram, the name of the cognate protein for each gene, and gene aliases (alternative names). The aliases in bold are gene names more commonly used by RAS researchers. Gene names, protein names, and gene aliases were obtained from BioGPS.
| Gene Name | Protein from BioGPS | Alternative Gene Names from BioGPS |
|---|---|---|
| AKT1 | RAC-alpha serine/threonine-protein kinase | AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA |
| AKT2 | RAC-beta serine/threonine-protein kinase | HIHGHH, PKBB, PKBBETA, PRKBB, RAC-BETA |
| AKT3 | RAC-gamma serine/threonine-protein kinase | MPPH, MPPH2, PKB-GAMMA, PKBG, PRKBG, RAC-PK-gamma, RAC-gamma, STK-2 |
| ALK | anaplastic lymphoma receptor tyrosine kinase; ALK tyrosine kinase receptor | CD246, NBLST3 |
| APAF1 | apoptotic peptidase activating factor 1 | APAF-1, CED4 |
| ARAF | serine/threonine-protein kinase A-Raf | A-RAF, ARAF1, PKS2, RAFA1 |
| ARHGAP35 | Rho GTPase-activating protein 35 | GRF-1, GRLF1, P190-A, P190A, p190ARhoGAP, p190RhoGAP |
| ARHGEF2 | Rho/Rac guanine nucleotide exchange factor (GEF) 2 | GEF, GEF-H1, GEFH1, LFP40, P40 |
| BARD1 | BRCA1 associated RING domain 1 | [none] |
| BRAF | serine/threonine-protein kinase B-raf | B-RAF1, BRAF1, NS7, RAFB1 |
| BRCA1 | breast cancer 1, early onset | BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 |
| BRCA2 | breast cancer 2, early onset | BRCC2, BROVCA2, FACD, FAD, FAD1, FANCD, FANCD1, GLM3, PNCA2 |
| BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | BACH1, FANCJ, OF |
| BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | BUB1A, BUB1L, hBUB1 |
| CASP3 | caspase 3, apoptosis-related cysteine peptidase | CPP32, CPP32B, SCA-1 |
| CASP7 | caspase 7, apoptosis-related cysteine peptidase | CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 |
| CASP8 | caspase 8, apoptosis-related cysteine peptidase | ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 |
| CBL | Cbl proto-oncogene, E3 ubiquitin protein ligase | C-CBL, CBL2, FRA11B, NSLL, RNF55 |
| CBLB | Cbl proto-oncogene B, E3 ubiquitin protein ligase | Cbl-b, RNF56 |
| CBLC | Cbl proto-oncogene C, E3 ubiquitin protein ligase | CBL-3, CBL-SL, RNF57 |
| CCNA1 | cyclin A1 | CT146 |
| CCNA2 | cyclin A2 | CCN1, CCNA |
| CCND1 | cyclin D1 | BCL1, D11S287E, PRAD1, U21B31 |
| CCND2 | cyclin D2 | KIAK0002, MPPH3 |
| CCND3 | cyclin D3 | [none] |
| CDC25A | cell division cycle 25A | CDC25A2 |
| CDC6 | cell division cycle 6 | cell division cycle 6 |
| CDK2 | cyclin-dependent kinase 2 | CDKN2, p33(CDK2) |
| CDK4 | cyclin-dependent kinase 4 | CMM3, PSK-J3 |
| CDK6 | cyclin-dependent kinase 6 | PLSTIRE |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 |
| CNKSR1 | connector enhancer of kinase suppressor of Ras 1 | CNK, CNK1 |
| CNKSR2 | connector enhancer of kinase suppressor of Ras 2 | CNK2, KSR2, MAGUIN |
| CYTH2 | cytohesin 2 | ARNO, CTS18, CTS18.1, PSCD2, PSCD2L, SEC7L, Sec7p-L, Sec7p-like |
| DAB2IP | DAB2 interacting protein | AF9Q34, AIP-1, AIP1, DIP1/2 |
| DHFR | dihydrofolate reductase | DHFRP1, DYR |
| DUSP1 | dual specificity phosphatase 1 | CL100, HVH1, MKP-1, MKP1, PTPN10 |
| DUSP2 | dual specificity phosphatase 2 | PAC-1, PAC1 |
| DUSP3 | dual specificity phosphatase 3 | VHR |
| DUSP4 | dual specificity phosphatase 4 | HVH2, MKP-2, MKP2, TYP |
| DUSP5 | dual specificity phosphatase 5 | DUSP, HVH3 |
| DUSP6 | dual specificity phosphatase 6 | HH19, MKP3, PYST1 |
| E2F1 | E2F transcription factor 1 | E2F-1, RBAP1, RBBP3, RBP3 |
| E2F2 | E2F transcription factor 2 | E2F-2 |
| E2F3 | E2F transcription factor 3 | E2F-3 |
| E2F7 | E2F transcription factor 7 | [none] |
| ECT2 | epithelial cell transforming 2 | ARHGEF31 |
| EGFR | epidermal growth factor receptor | ERBB, ERBB1, HER1, PIG61, mENA |
| EIF4EBP1 | eukaryotic translation initiation factor 4E-binding protein 1 | 4E-BP1, 4EBP1, BP-1, PHAS-I |
| EIF4EBP2 | eukaryotic translation initiation factor 4E binding protein 2 | 4EBP2, PHASII |
| EIF4EBP3 | eukaryotic translation initiation factor 4E binding protein 3 | 4E-BP3, 4EBP3 |
| ELK1 | ELK1, member of ETS oncogene family | [none] |
| ERBB2 | receptor tyrosine-protein kinase erbB-2 | CD340, HER-2, HER-2/neu, HER2, MLN 19, NEU, NGL, TKR1 |
| ERF | Ets2 repressor factor | CRS4, PE-2, PE2 |
| ETS1 | v-ets avian erythroblastosis virus E26 oncogene homolog 1 | ETS-1, EWSR2, p54 |
| ETS2 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 | ETS2IT1 |
| EXOC1 | Exocyst complex component 1 | BM-102, SEC3, SEC3L1, SEC3P |
| EXOC2 | exocyst complex component 2 | SEC5, SEC5L1, Sec5p |
| EXOC3 | exocyst complex component 3 | SEC6, SEC6L1, Sec6p |
| EXOC4 | exocyst complex component 4 | SEC8, SEC8L1, Sec8p |
| EXOC5 | exocyst complex component 5 | HSEC10, PRO1912, SEC10, SEC10L1, SEC10P |
| EXOC6 | exocyst complex component 6 | EXOC6A, SEC15, SEC15L, SEC15L1, SEC15L3, Sec15p |
| EXOC7 | exocyst complex component 7 | 2-5-3p, EX070, EXO70, EXOC1, Exo70p, YJL085W |
| EXOC8 | exocyst complex component 8 | EXO84, Exo84p, SEC84 |
| FANCA | Fanconi anemia, complementation group A | FA, FA-H, FA1, FAA, FACA, FAH, FANCH |
| FANCC | Fanconi anemia, complementation group C | FA3, FAC, FACC |
| FGFR1 | fibroblast growth factor receptor 1 | BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 |
| FGFR2 | fibroblast growth factor receptor 2 | BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 |
| FGFR3 | fibroblast growth factor receptor 3 | CH, CD333, CEK2, HSFGFR3EX, JTK4 |
| FGFR4 | fibroblast growth factor receptor 4 | CD334, JTK2, TKF |
| FLT3 | fms-related tyrosine kinase 3 | CD135, FLK-2, FLK2, STK1 |
| FNTA | farnesyltransferase, CAAX box, alpha | FPTA, PGGT1A, PTAR2 |
| FNTB | farnesyltransferase, CAAX box, beta | FPTB |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | AP-1, C-FOS, p55 |
| FOSL1 | FOS-like antigen 1 | FRA, FRA1, fra-1 |
| GRB10 | growth factor receptor-bound protein 10 | GRB-IR, Grb-10, IRBP, MEG1, RSS |
| GRB2 | growth factor receptor-bound protein 2 | ASH, EGFRBP-GRB2, Grb3-3, MST084, MSTP084, NCKAP2 |
| HRAS | GTPase HRas | C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV, HRAS1, RASH1, p21ras |
| ICMT | isoprenylcysteine carboxyl methyltransferase | HSTE14, MST098, MSTP098, PCCMT, PCMT, PPMT |
| INPP4B | inositol polyphosphate-4-phosphatase, type II, 105kDa | [none] |
| INSR | insulin receptor | CD220, HHF5 |
| INSRR | insulin receptor-related receptor | IRR |
| IRS1 | insulin receptor substrate 1 | HIRS-1 |
| IRS2 | insulin receptor substrate 2 | IRS-2 |
| JUN | Jun proto-oncogene | AP-1, AP1, c-Jun |
| KRAS | GTPase KRas | C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2 |
| KSR1 | kinase suppressor of Ras 1 | KSR, RSU2 |
| KSR2 | kinase suppressor of Ras 2 | [none] |
| MAP2K1 | dual specificity mitogen-activated protein kinase kinase 1 | CFC3, MAPKK1, MEK1, MKK1, PRKMK1 |
| MAP2K2 | dual specificity mitogen-activated protein kinase kinase 2 | CFC4, MAPKK2, MEK2, MKK2, PRKMK2 |
| MAPK1 | mitogen-activated protein kinase 1 | ERK, ERK-2, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk, p42-MAPK |
| MAPK3 | mitogen-activated protein kinase 3 | ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK |
| MCM3 | minichromosome maintenance complex component 3 | HCC5, P1-MCM3, P1.h, RLFB |
| MCM4 | minichromosome maintenance complex component 4 | CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 |
| MCM5 | minichromosome maintenance complex component 5 | CDC46, P1-CDC46 |
| MCM6 | minichromosome maintenance complex component 6 | MCG40308, Mis5, P105MCM |
| MCM7 | minichromosome maintenance complex component 7 | CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146, PPP1R104 |
| MDM2 | MDM2 proto-oncogene, E3 ubiquitin protein ligase | ACTFS, HDMX, hdm2 |
| MET | MET proto-oncogene, receptor tyrosine kinase; hepatocyte growth factor receptor | AUTS9, HGFR, RCCP2, c-Met |
| MLST8 | MTOR associated protein, LST8 homolog (S. cerevisiae) | GBL, GbetaL, LST8, POP3, WAT1 |
| MRAS | muscle RAS oncogene homolog | M-RAs, R-RAS3, RRAS3 |
| MTOR | mechanistic target of rapamycin (serine/threonine kinase) | FRAP, FRAP1, FRAP2, RAFT1, RAPT1 |
| MYB | v-myb avian myeloblastosis viral oncogene homolog | Cmyb, c-myb, c-myb_CDS, efg |
| MYC | v-myc avian myelocytomatosis viral oncogene homolog | MRTL, MYCC, bHLHe39, c-Myc |
| NF1 | neurofibromin | NFNS, VRNF, WSS |
| NFE2L2 | nuclear factor, erythroid 2-like 2 | NRF2 |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 |
| NOX1 | NADPH oxidase 1 | GP91-2, MOX1, NOH-1, NOH1 |
| NRAS | GTPase NRas | ALPS4, CMNS, N-ras, NCMS, NRAS1, NS6 |
| PAK1 | p21 protein (Cdc42/Rac)-activated kinase 1 | PAKalpha |
| PAK2 | p21 protein (Cdc42/Rac)-activated kinase 2 | PAK65, PAKgamma |
| PAK3 | p21 protein (Cdc42/Rac)-activated kinase 3 | CDKN1A, MRX30, MRX47, OPHN3, PAK3beta, bPAK, hPAK3 |
| PAK4 | p21 protein (Cdc42/Rac)-activated kinase 4 | [none] |
| PDGFRA | platelet-derived growth factor receptor alpha | CD140A, PDGFR-2, PDGFR2, RHEPDGFRA |
| PDGFRB | platelet-derived growth factor receptor beta | CD140B, IBGC4, IMF1, JTK12, PDGFR, PDGFR-1, PDGFR1 |
| PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | PDK1, PDPK2, PRO0461 |
| PEA15 | phosphoprotein enriched in astrocytes 15 | HMAT1, HUMMAT1H, MAT1, MAT1H, PEA-15, PED |
| PEBP1 | phosphatidylethanolamine binding protein 1 | HCNP, HCNPpp, HEL-210, HEL-S-34, PBP, PEBP, PEBP-1, RKIP |
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | CLOVE, CWS5, MCAP, MCM, MCMTC, PI3K, p110-alpha |
| PIK3CD | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta | APDS, IMD14, P110DELTA, PI3K, p110D |
| PIK3CG | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma | PI3CG, PI3K, PI3Kgamma, PIK3, p110gamma, p120-PI3K |
| PIK3R1 | phosphatidylinositol 3-kinase regulatory subunit alpha | AGM7, GRB1, p85, p85-ALPHA |
| PIK3R2 | phosphatidylinositol 3-kinase regulatory subunit beta | MPPH, MPPH1, P85B, p85, p85-BETA |
| PIK3R3 | phosphatidylinositol 3-kinase regulatory subunit gamma | p55, p55-GAMMA |
| PIK3R5 | phosphoinositide 3-kinase regulatory subunit 5 | F730038I15Rik, FOAP-2, P101-PI3K, p101 |
| PIK3R6 | phosphoinositide 3-kinase regulatory subunit 6 | C17orf38, HsT41028, p84 PIKAP, p87(PIKAP), p87PIKAP |
| PIN1 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | DOD, UBL5 |
| PLCE1 | phospholipase C, epsilon 1 | NPHS3, PLCE, PPLC |
| PLXNB1 | plexin B1 | PLEXIN-B1, PLXN5, SEP |
| PPP1CA | protein phosphatase 1, catalytic subunit, alpha isozyme | PP-1A, PP1A, PP1alpha, PPP1A |
| PREX2 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 | 6230420N16Rik, DEP.2, DEPDC2, P-REX2, PPP1R129 |
| PRKAA1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | AMPK, AMPKa1 |
| PRKAA2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | AMPK, AMPK2, AMPKa2, PRKAA |
| PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1 | AMPK, HAMPKb |
| PRKAB2 | 5'-AMP-activated protein kinase subunit beta-2 | [none] |
| PRKAG1 | protein kinase, AMP-activated, gamma 1 non-catalytic subunit | AMPKG |
| PRKAG2 | protein kinase, AMP-activated, gamma 2 non-catalytic subunit | AAKG, AAKG2, CMH6, H91620p, WPWS |
| PRKAG3 | protein kinase, AMP-activated, gamma 3 non-catalytic subunit | AMPKG3 |
| PTEN | phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 |
| PTPN11 | protein tyrosine phosphatase, non-receptor type 11 | BPTP3, CFC, NS1, PTP-1D, PTP2C, SH-PTP2, SH-PTP3, SHP2 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Rac-1, TC-25, p21-Rac1 |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | EN-7, Gx, HSPC022, p21-Rac2 |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | [none] |
| RAD52 | RAD52 homolog (S. cerevisiae) | [none] |
| RAF1 | RAF proto-oncogene serine/threonine-protein kinase | CMD1NN, CRAF, NS5, Raf-1, c-Raf |
| RALA | Ras-related protein Ral-A | RAL |
| RALB | Ras-related protein Ral-B | [none] |
| RALBP1 | RalA binding protein 1 | RIP1, RLIP1, RLIP76 |
| RALGAPA1 | Ral GTPase activating protein, alpha subunit 1 (catalytic) | GARNL1, GRIPE, RalGAPalpha1, TULIP1, p240 |
| RALGAPA2 | Ral GTPase activating protein, alpha subunit 2 (catalytic) | AS250, C20orf74, bA287B20.1, dJ1049G11, dJ1049G11.4, p220 |
| RALGDS | Ral guanine nucleotide dissociation stimulator | RGDS, RGF, RalGEF |
| RAPGEF1 | Rap guanine nucleotide exchange factor (GEF) 1 | C3G, GRF2 |
| RAPGEF2 | Rap guanine nucleotide exchange factor (GEF) 2 | CNrasGEF, NRAPGEP, PDZ-GEF1, PDZGEF1, RA-GEF, RA-GEF-1, Rap-GEP, nRap GEP |
| RASA1 | Ras GTPase-activating protein 1 | CM-AVM, CMAVM, GAP, PKWS, RASA, RASGAP, p120GAP, p120RASGAP |
| RASA2 | Ras GTPase-activating protein 2 | GAP1M |
| RASA3 | Ras GTPase-activating protein 3 | GAP1IP4BP, GAPIII |
| RASAL1 | RasGAP-activating-like protein 1 | RASAL |
| RASAL2 | Ras GTPase-activating protein nGAP | NGAP |
| RASAL3 | RAS protein activator like-3 | [none] |
| RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 | CDC25, CDC25L, GNRP, GRF1, GRF55, H-GRF55, PP13187, ras-GRF1 |
| RASGRF2 | Ras protein-specific guanine nucleotide-releasing factor 2 | GRF2, RAS-GRF2 |
| RASGRP1 | RAS guanyl-releasing protein 1 | CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 |
| RASGRP2 | RAS guanyl-releasing protein 2 | CALDAG-GEFI, CDC25L |
| RASGRP3 | Ras guanyl-releasing protein 3 | GRP3, KIAA0846 |
| RASGRP4 | RAS guanyl-releasing protein 4 | [none] |
| RASSF1 | Ras association (RalGDS/AF-6) domain family member 1 | 123F2, NORE2A, RASSF1A, RDA32, REH3P21 |
| RASSF10 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 10 | [none] |
| RASSF2 | Ras association (RalGDS/AF-6) domain family member 2 | CENP-34, RASFADIN |
| RASSF3 | Ras association (RalGDS/AF-6) domain family member 3 | RASSF5 |
| RASSF4 | Ras association (RalGDS/AF-6) domain family member 4 | AD037 |
| RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | Maxp1, NORE1, NORE1A, NORE1B, RAPL, RASSF3 |
| RASSF6 | Ras association (RalGDS/AF-6) domain family member 6 | [none] |
| RASSF7 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 | C11orf13, HRAS1, HRC1 |
| RASSF8 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 | C12orf2, HOJ1 |
| RASSF9 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 | P-CIP1, PAMCI, PCIP1 |
| RB1 | Retinoblastoma 1 | OSRC, PPP1R130, RB, p105-Rb, pRb, pp110 |
| RCE1 | Ras converting CAAX endopeptidase 1 | FACE2, RCE1A, RCE1B |
| RGL1 | ral guanine nucleotide dissociation stimulator-like 1 | RGL |
| RGL2 | ral guanine nucleotide dissociation stimulator-like 2 | HKE1.5, KE1.5, RAB2L |
| RGL3 | ral guanine nucleotide dissociation stimulator-like 3 | [none] |
| RHEB | Ras Homolog Enriched In Brain, GTP-binding protein Rheb | RHEB2 |
| RHOA | Ras homolog family member A | ARH12, ARHA, RHO12, RHOH12 |
| RHOB | Ras homolog family member B | ARH6, ARHB, MST081, MSTP081, RHOH6 |
| RHOC | Ras homolog family member C | ARH9, ARHC, H9, RHOH9 |
| ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | P160ROCK, ROCK-I |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | ROCK-II |
| ROS1 | ROS proto-oncogene 1 , receptor tyrosine kinase | MCF3, ROS, c-ros-1 |
| RPS6KA1 | ribosomal protein S6 kinase alpha-1 | MAPKAPK1A, RSK1 |
| RPS6KA2 | ribosomal protein S6 kinase alpha-2 | HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-alpha2, p90-RSK3, pp90RSK3 |
| RPS6KA3 | ribosomal protein S6 kinase alpha-3 | CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 |
| RPS6KA6 | ribosomal protein S6 kinase alpha-6 | PP90RSK4, RSK4 |
| RPS6KB1 | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | PS6K, S6K, S6K-beta-1, S6K1, STK14A, p70 S6KA, p70(S6K)-alpha, p70-S6K, p70-alpha |
| RPS6KB2 | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | KLS, P70-beta, P70-beta-1, P70-beta-2, S6K-beta2, S6K2, SRK, STK14B, p70(S6K)-beta, p70S6Kb |
| RPTOR | regulatory associated protein of MTOR, complex 1 | KOG1, Mip1 |
| SAV1 | salvador family WW domain containing protein 1 | SAV, WW45, WWP4 |
| SCRIB | scribbled planar cell polarity protein | CRIB1, SCRB1, SCRIB1, Vartul |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | SHC, SHCA |
| SHC2 | SHC (Src homology 2 domain containing) transforming protein 2 | SCK, SHCB, SLI |
| SHC3 | SHC (Src homology 2 domain containing) transforming protein 3 | N-Shc, NSHC, RAI, SHCC |
| SHC4 | SHC (Src homology 2 domain containing) transforming protein 4 | RaLP, SHCD |
| SHOC2 | Soc-2 suppressor of clear homolog (C. elegans) | SIAA0862, SOC2, SUR8 |
| SOS1 | Son of sevenless homolog 1 | GF1, GGF1, GINGF, HGF, NS4 |
| SOS2 | Son of sevenless homolog 2 | [none] |
| SPRED1 | Sprouty-related, EVH1 domain-containing protein 1 | NFLS, PPP1R147, hSpred1, spred-1 |
| SPRED2 | Sprouty-related, EVH1 domain-containing protein 2 | Spred-2 |
| SPRED3 | Sprouty-related, EVH1 domain-containing protein 3 | Eve-3, spred-3 |
| SPRY1 | protein sprouty homolog 1 | hSPRY1 |
| SPRY2 | protein sprouty homolog 2 | hSPRY2 |
| SPRY3 | protein sprouty homolog 3 | HSPRY3, spry-3 |
| SPRY4 | protein sprouty homolog 4 | HH17 |
| STK11 | serine/threonine-protein kinase STK11 | LKB1, PJS, hLKB1 |
| STK3 | serine/threonine kinase 3 | KRS1, MST2 |
| STK4 | serine/threonine kinase 4 | KRS2, MST1, TIIAC, YSK3 |
| TBK1 | TANK-binding kinase 1 | NAK, T2K |
| TFDP1 | transcription factor Dp-1 | DP1, DRTF1, Dp-1 |
| TFDP2 | transcription factor Dp-2 (E2F dimerization partner 2) | DP2 |
| TIAM1 | T-lymphoma invasion and metastasis-inducing protein 1 | [none] |
| TIAM2 | T-lymphoma invasion and metastasis-inducing protein 2 | STEF, TIAM-2 |
| TK1 | thymidine kinase 1, soluble | TK2 |
| TP53 | tumor protein p53 | BCC7, LFS1, P53, TRP53 |
| TP73 | tumor protein p73 | P73 |
| TSC1 | Tuberous Sclerosis 1, Hamartin | LAM, TSC |
| TSC2 | Tuberous Sclerosis 2, Tuberin | LAM, PPP1R160, TSC4 |
| TYMS | thymidylate synthetase | HST422, TMS, TS |
| UNG | uracil-DNA glycosylase | DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 |
| VAV1 | vav 1 guanine nucleotide exchange factor | VAV |
| YAP1 | Yes-associated protein 1 | COB1, YAP, YAP2, YAP65, YKI |