OCG’s Data Coordinating Center: Facilitating the Sharing of Data Generated by the Office’s Initiatives
, by Yiwen He, Ph.D., and DCC staff
Today, cancer research studies by different research groups produce vast amounts of data in widely varying formats. In order to continuously improve our understanding of cancer mechanisms which in turn could improve patient outcomes, it is important for these data to be effectively analyzed, interpreted, and utilized by other studies according to the FAIR (findable, accessible, interoperable, and reusable) principles throughout the life of each study1. The Office of Cancer Genomics’ (OCG) Data Coordinating Center (DCC) is responsible for managing the flow of data within each of OCG’s programs: Therapeutically Applicable Research to Generate Effective Treatments (TARGET), Cancer Genome Characterization Initiative (CGCI), Cancer Target Discovery and Development (CTD²) Network, and Human Cancer Models Initiative (HCMI). The DCC’s major goals are to collect the multiple datasets generated within each program, check submitted data files for quality (QC), assist in harmonizing the data into standard formats, and perform or facilitate distribution of the data to other centralized repositories such as the National Cancer Institute's (NCI) Genomics Data Commons (GDC) and National Center for Biotechnology Information’s (NCBI) Sequence Read Archive (SRA).
While most raw sequencing data such as FASTQ2 and BAM3 files are submitted by sequencing centers directly to GDC (previously to SRA), clinical, biospecimen, and higher level (analyzed) data are submitted to the DCC. The DCC then submits the clinical and biospecimen data to the other repositories mentioned above, and also makes each project’s data files available for download from the DCC’s servers.
Project team members work directly with the DCC to obtain user accounts in order to submit (upload) data to or download data from the DCC’s servers via a secure file transfer protocol (sFTP). The DCC stores the data for collaborators to use while the studies are in progress, and depending on the dataset, releases the data to the public or to dbGaP-approved, controlled-access users once a related research paper has been accepted for publication. The Guide to Accessing Data provides a visual and interactive overview on how to obtain approval to view controlled-access datasets. Once datasets are released at the DCC, OCG provides several tools to help users navigate and understand the data associated with each study.
Navigating Through CGCI and TARGET Projects at the DCC
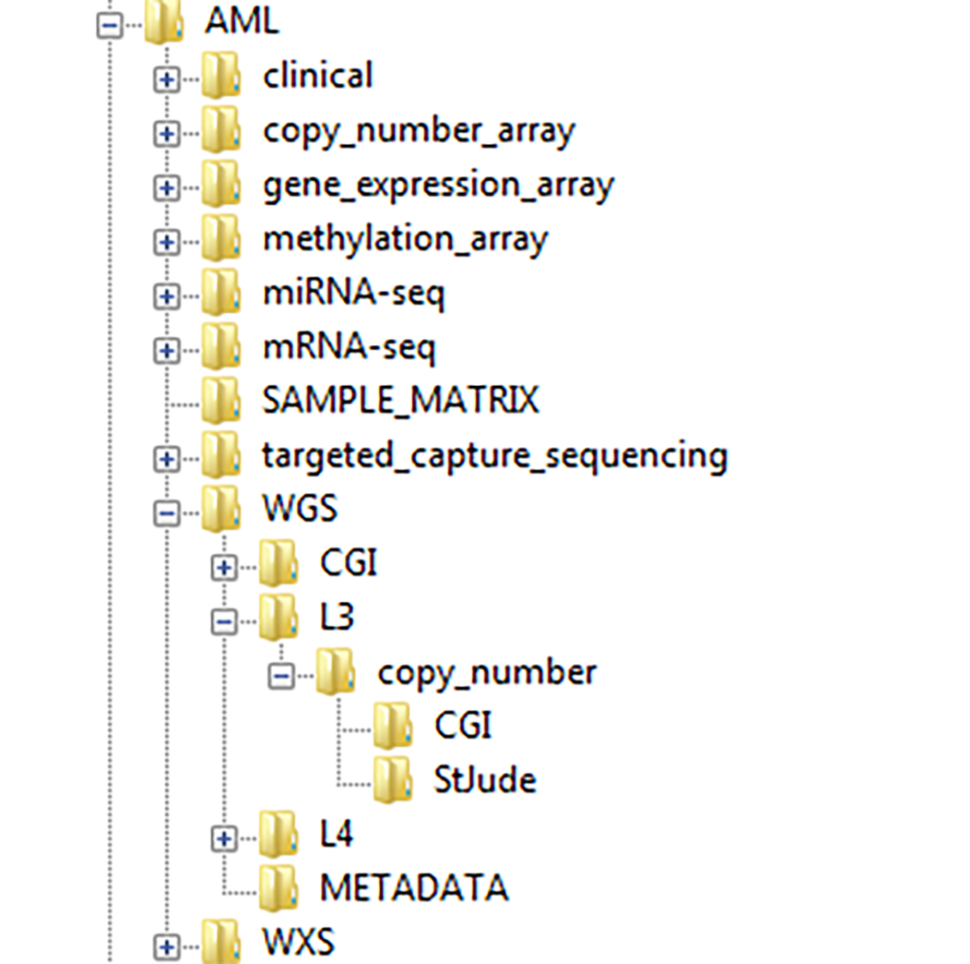
The CGCI Data Matrix and the TARGET Data Matrix provide centralized starting points to the data types available for each project, and to navigate those datasets that are stored at the DCC. The data matrices direct users to specific locations from which it is possible to download the dataset of interest. In general, data stored at the DCC is organized within directories that correspond to the types of analysis, levels of data, and the names of centers that submitted a particular type of data when there are multiple data-submitting centers.
Users can download the data via links provided in the data matrices or via Unix by using commands such as wget and curl. Description on how to use the data can be found on the Using CGCI Data and Using TARGET Data webpages. The DCC also provides users with more in-depth information on data availability for each project within TARGET and CGCI in the form of Sample Matrices.
CGCI and TARGET Sample Matrices
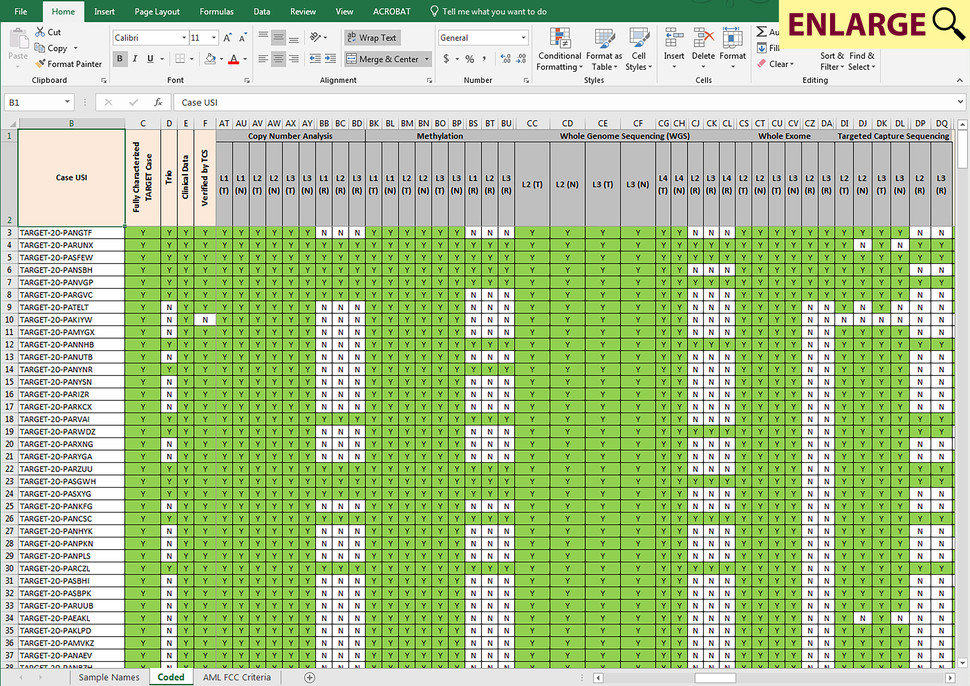
Sample matrices, available in each project’s SAMPLE_MATRIX download directory at the DCC, specify the type of data available for each patient/case for each cancer type within the CGCI and TARGET programs. Depending on the study, each row of a sample matrix may correspond to either a single case or sample, and the columns within a row show availability of data from different analyses (e.g. either chip-based or sequence based characterization of RNA, genome, exome, targeted capture or epigenome), as well as data levels for the corresponding case/sample. A “Comments” column provided at the end of each row may contain additional information that is important to note for a particular case/sample.
CGCI and TARGET Analysis Metadata
Metadata files, including MAGE-TAB-formatted SDRF and IDF files, map cases within a study to related data files produced by the project. These files can be found in the METADATA directory of each type of analysis.
CTD² Data Resources
CTD² is a “community resource project”, meaning members of the Network are required to release data to the research community. The release of CTD² data to the scientific community is intended to maximize the impact of the findings. Data generated by the Network can be accessed through two resources: CTD² Data Portal and CTD² Dashboard.
CTD² Data Portal
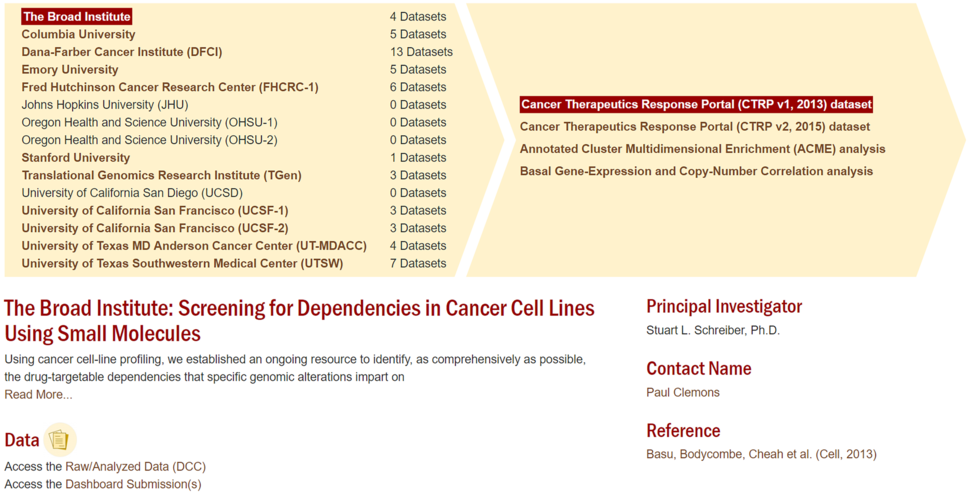
The CTD² Data Portal is an open-access resource which hosts raw/analyzed primary data generated from different types of experimental and computational approaches. The data can be sorted by Center or the method used to generate the data. Along with the Data Harmonization Informatics Portal team (comprising at least one member from each Center), the DCC is responsible for quality assurance and usability of the data submissions. The DCC currently supports data from 15 Centers. In the near future, data from Network Centers’ collaborations will be listed under “Cross-Center Projects” and will be noted with an asterisk. Data stored at the DCC can be accessed through the “Raw/Analyzed Data” link on each project page on the Data Portal. Sequence data that are categorized by the Centers’ Institutional Review Boards as “open access” are stored at NCBI’s Gene Expression Omnibus (GEO).
CTD² Dashboard Data
The CTD² Dashboard is an open-access web interface with observations compiled from the data generated by various types of biological and analytical approaches by the CTD² Centers. The DCC is responsible for maintaining the website and creating monthly releases of the Dashboard to update the observations when the Centers submit them. Some projects on the Data Portal have a link to the corresponding Dashboard submissions.
Conclusion
In summary, the DCC provides bioinformatic support for the OCG cancer research programs. The DCC supports OCG by receiving and formatting genomics data, transferring data to GDC, maintaining web applications, and coordinating with NIH’s Center for Information Technology regarding security, storage space, and web hosting. The DCC plays a key role in ensuring that the standardized genomic data from the OCG programs can be systematically accessed by the cancer research community and that the process adheres to the FAIR principles.
References
- Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data. 2016 Mar 15;3:160018. (PMID: 26978244)
- Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research. 2010 Apr;38(6):1767-71. (PMID: 20015970)
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 Aug 15;25(16):2078-9. (PMID: 19505943)