Introducing the Genomics Data Commons
, by Nadia Jaber
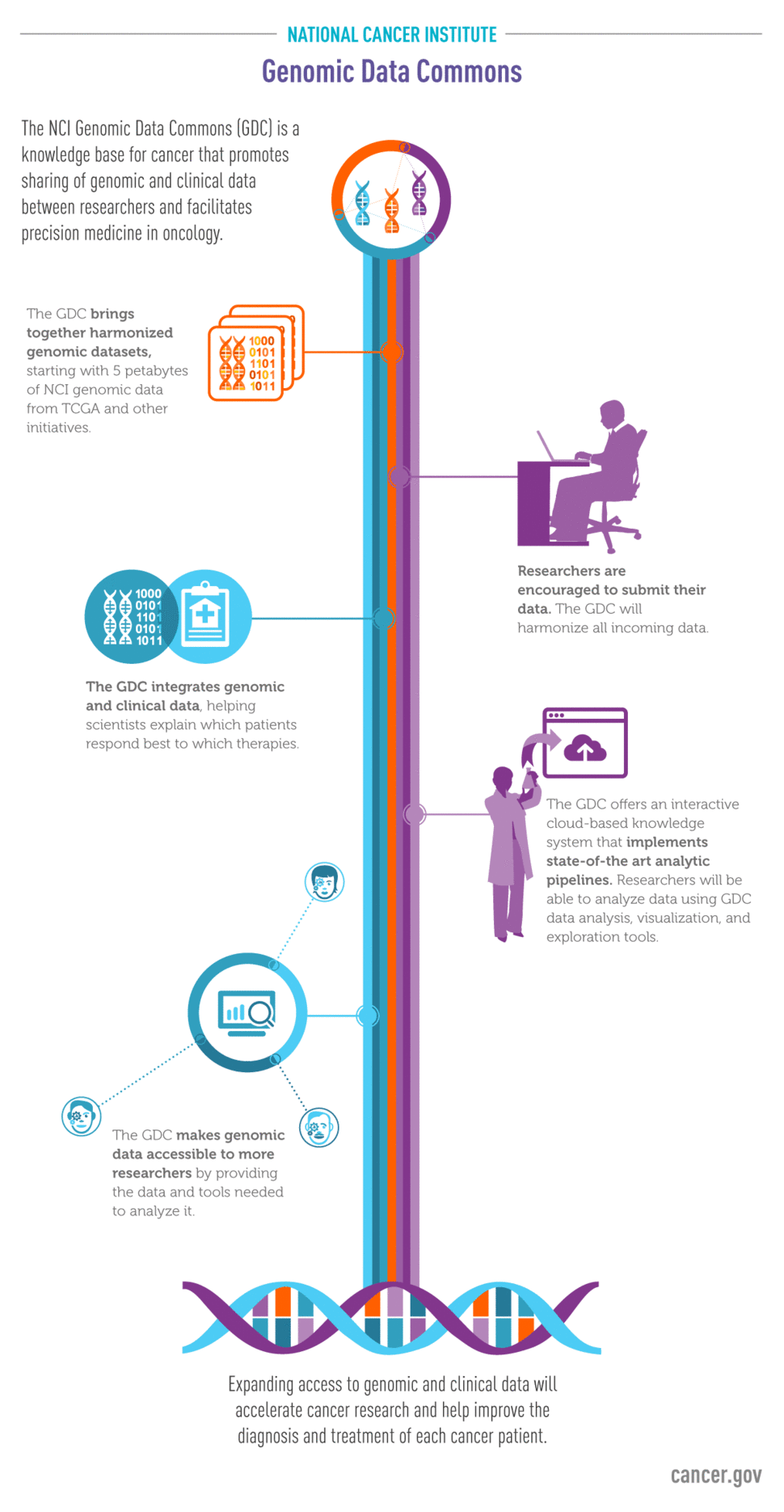
As genomics studies progress and datasets become larger and more complex, the ability of the research community to access and analyze genomic data is hindered by several limitations, including the size of data files, cost of storage, and difficulty accessing various portals. To address these challenges, the Center for Cancer Genomics launched the Genomics Data Commons (GDC): a unified data sharing platform which enables data sharing across the entire cancer research community, to ultimately support precision medicine in oncology.
As a secure data storage network, the GDC provides investigators with a single portal to access genomic characterization datasets including The Cancer Genome Atlas (TCGA) and Therapeutically Applicable Research to Generate Effective Treatments (TARGET). In the future, other datasets, including the Cancer Genome Characterization Initiative (CGCI), the Cancer Cell Line Encyclopedia (CCLE), and the Human Cancer Models Initiative (HCMI) will be added. In addition, the molecular information company Foundation Medicine, Inc., which has already pledged to add data from 18,000 adult cancer patients.
For the community of researchers that contribute to and follow OCG research programs, the launch of the GDC means that data from TARGET and CGCI (but not CTD2) will be accessible through the GDC in addition to their designated data matrices. The raw data files used to produce the analyzed data accessible from the OCG matrices are the same as those stored in the GDC. The GDC will use 17 different analytical pipelines to analyze the TARGET data and will map the sequences to the latest version of the human genome reference sequence. Therefore, re-aligned and re-analyzed data in the GDC may be different from those accessible through the OCG data matrices. Additionally, the data access sites differ in their user interfaces and interactive applications. In the OCG matrices, data are separated by program and further delineated by project and tissue type. In the default setting for the GDC data are not separated by any factors, so all programs and tissue types are displayed. However, data can be sorted by program (such as TARGET), primary site (such as kidney), disease type (such as high-risk Wilms Tumor), data category (like transcriptome profiling), or experimental strategy (like whole exome sequencing). For example, users can view, download, and analyze TARGET’s dataset on its own, or in conjunction with other datasets. It should be noted that although the datasets are intermingled, users will need a separate data use certification (DUC) to access each program’s data.
Another advantage of the GDC is that data are harmonized (meaning uniformly analyzed) which enables the direct comparison and analysis of datasets from different sources in ways that were not possible before. Data harmonization allows investigators to carry out analyses on cases from multiple studies, thereby enhancing statistical power and increasing the depth of investigation. This is especially important for rare and understudied cancers, such as those studied by TARGET and CGCI.
The GDC also holds clinical information associated with the molecular data, and a long-term goal of the GDC is for physicians to use it as a tool for precision oncology. In addition, GDC users have the ability to upload genomic data, increasing the breadth of data and allowing for more comparisons. Alternatively, a provider could possibly determine the best course of treatment to match the specific genetic vulnerabilities of the patient’s tumor by looking at the associated clinical information, such as treatment regimen and outcome, from other patients with the same alterations. A cancer “knowledge bank” containing both genomic and clinical data will be a critical component of precision oncology strategies, and the GDC has the potential to be one component.
The GDC will continue to grow with data, tools, and resources, and has the potential to transform the use of OCG and other genomic datasets.
For more details about the GDC:
- View the GDC website
- Launch the GDC Data Portal
- Read the NIH press release
- Read the NCI News Note
- Check out the GDC fact sheet
- Watch an introductory video featuring CCG Director Dr. Louis Staudt
- Visit the GDC About the Data page
- Read about the addition of data from Foundation Medicine, Inc.
- For questions and feedback, email the GDC support team
- Follow the GDC on Twitter: @NCIGDC_Updates
- For help accessing and submitting data, visit GDC Support Resources