Genetics of Colorectal Cancer (PDQ®)–Health Professional Version
Executive Summary
This executive summary reviews the topics covered in the PDQ summary on the genetics of colorectal cancer (CRC), with hyperlinks to detailed sections below that describe the evidence on each topic.
-
Inheritance and Risk
Factors suggestive of a genetic contribution to CRC include the following: (1) a strong family history of CRC and/or polyps; (2) multiple primary cancers in a patient with CRC; (3) the existence of other cancers within the kindred consistent with known syndromes causing an inherited risk of CRC, such as endometrial cancer; and (4) early age at diagnosis of CRC. Hereditary CRC is most commonly inherited in an autosomal dominant pattern, although two syndromes are inherited in an autosomal recessive pattern (MUTYH-associated polyposis and NTHL1).
At least three validated computer models are available to estimate the probability that an individual affected with cancer carries a pathogenic variant in a mismatch repair (MMR) gene associated with Lynch syndrome, the most common inherited CRC syndrome. These include the MMRpro, MMRpredict, and PREMM5 (PREdiction Model for gene Mutations) prediction models. Individuals with a quantified risk of 2.5% or greater on PREMM5 or 5% or greater on MMRpro and MMRpredict are recommended for genetic evaluation referral and testing.
-
Associated Genes and Syndromes
Hereditary CRC has two well-described forms: (1) polyposis (including familial adenomatous polyposis [FAP] and attenuated FAP [AFAP], which are caused by pathogenic variants in the APC gene; and MUTYH-associated polyposis, which is caused by pathogenic variants in the MUTYH gene); and (2) Lynch syndrome (often referred to as hereditary nonpolyposis colorectal cancer), which is caused by germline pathogenic variants in DNA MMR genes (MLH1, MSH2, MSH6, and PMS2) and EPCAM. Other CRC syndromes and their associated genes include oligopolyposis (POLE, POLD1), NTHL1, juvenile polyposis syndrome (BMPR1A, SMAD4), Cowden syndrome (PTEN), and Peutz-Jeghers syndrome (STK11). Many of these syndromes are also associated with extracolonic cancers and other manifestations. Serrated polyposis syndrome, which is characterized by the appearance of hyperplastic polyps, appears to have a familial component, but the genetic basis remains unknown. The natural history of some of these syndromes is still being described. Many other families exhibit aggregation of CRC and/or adenomas, but with no apparent association with an identifiable hereditary syndrome, and they are known collectively as familial CRC. In addition, most individuals with CRC diagnosed before age 50 years and without a family history of cancer do not have a pathogenic variant associated with an inherited cancer syndrome.
Genome-wide searches are showing promise in identifying common, low-penetrance susceptibility alleles for many complex diseases, including CRCs, but the clinical utility of these findings remains uncertain.
-
Clinical Management
It is becoming the standard of care at many centers that all individuals with newly diagnosed CRC are evaluated for Lynch syndrome through molecular diagnostic tumor testing assessing MMR deficiency. A universal screening approach to tumor testing is supported, in which all CRC cases are evaluated regardless of age at diagnosis or fulfillment of existing clinical criteria for Lynch syndrome. A more cost-effective approach has been reported whereby all patients aged 70 years or younger with CRC and older patients who meet the revised Bethesda guidelines are tested for Lynch syndrome. Tumor evaluation often begins with immunohistochemistry testing for the expression of the MMR proteins associated with Lynch syndrome or microsatellite instability (MSI) testing, BRAF testing, and MLH1 hypermethylation analyses.
Colonoscopy for CRC screening and surveillance is commonly performed in individuals with hereditary CRC syndromes and has been associated with improved survival outcomes. For example, surveillance of Lynch syndrome patients with colonoscopy every 1 to 2 years, and in one study up to 3 years, has been shown to reduce CRC incidence and mortality. Extracolonic surveillance is also a mainstay for some hereditary CRC syndromes depending on the other cancers associated with the syndrome. For example, regular endoscopic surveillance of the duodenum in FAP patients has been shown to improve survival.
Prophylactic surgery (colectomy) has also been shown to improve survival in patients with FAP. The timing and extent of risk-reducing surgery usually depends on the number of polyps, their size, histology, and symptomatology. For patients with Lynch syndrome and a diagnosis of CRC, extended resection is associated with fewer metachronous CRCs and additional surgical procedures for colorectal neoplasia than in patients who undergo segmental resection for CRC. The surgical decision must consider the age of the patient, comorbidities, clinical stage of the tumor, sphincter function, and the patient’s wishes.
Chemopreventive agents have also been studied in the management of FAP and Lynch syndrome. In FAP patients, celecoxib and sulindac have been associated with a decrease in polyp size and number. A double-blind, randomized, controlled trial evaluating the efficacy of sulindac plus an epidermal growth factor receptor inhibitor, erlotinib, versus placebo in FAP or AFAP patients with duodenal polyps suggested that erlotinib has the potential to inhibit duodenal polyps in FAP patients. An ongoing trial will determine whether lower doses of erlotinib alone will significantly reduce duodenal polyp burden. Aspirin use (600 mg daily) was shown to have a preventive effect on cancer incidence in Lynch syndrome patients in a large randomized trial; lower doses are being examined in an ongoing study.
Novel therapies that stimulate the immune system have been evaluated in MMR-deficient tumors, including those related to Lynch syndrome. The dense immune infiltration and cytokine-rich environment in MMR-deficient tumors may improve clinical outcomes. A critical pathway responsible for mediating tumor-induced immune suppression is the programmed cell death-1 (PD-1)–mediated checkpoint pathway. Two phase 2 studies using anti–PD-1 immune checkpoint inhibitors (pembrolizumab and nivolumab) demonstrated favorable outcomes, including progression-free survival, radiographic response rates, and disease control rates in metastatic CRC with MMR deficiency and MSI that had progressed on prior cytotoxic chemotherapy. Pembrolizumab has shown similar benefit in other noncolorectal cancers with MMR deficiency and MSI, but not in tumors that are microsatellite stable.
-
Psychosocial and Behavioral Issues
Psychosocial factors influence decisions about genetic testing for inherited cancer risk and risk-management strategies. Uptake of genetic counseling and genetic testing for Lynch syndrome and FAP varies widely across studies. Factors that have been associated with genetic counseling and testing uptake in Lynch syndrome families include having children, the number of affected relatives, perceived risk of developing CRC, and frequency of thoughts about CRC. Psychological studies have shown low levels of distress, particularly in the long term, after genetic testing for Lynch syndrome in both carriers and noncarriers. However, other studies have demonstrated the possibility of increased distress following genetic testing for FAP. Colon and gynecologic cancer screening rates have been shown to increase or be maintained among carriers of MMR pathogenic variants within the year after disclosure of results, while screening rates decrease among noncarriers. The latter is expected as the screening recommendations for unaffected individuals are those that apply to the general population. Studies measuring quality-of-life variables in FAP patients show normal-range results; however, these studies suggest that risk-reducing surgery for FAP may have negative quality-of-life effects for at least some proportion of those affected. Patients' communication with their family members about an inherited risk of CRC is complex; gender, age, and the degree of relatedness are some elements that affect disclosure of this information. Research is ongoing to better understand and address psychosocial and behavioral issues in high-risk families.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in both men and women.
Estimated new cases and deaths from CRC in 2024 in the United States:[1]
- New cases: 152,810.
- Deaths: 53,010.
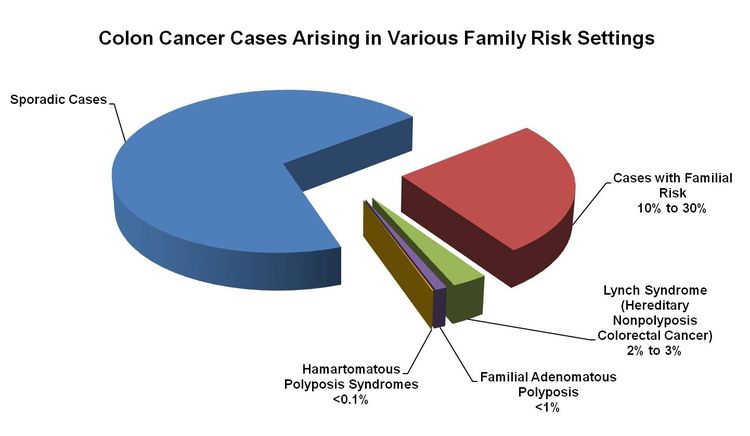
About 75% of patients with CRC have sporadic disease with no apparent evidence of having inherited the disorder. The remaining 10% to 30% of patients have a family history of CRC that suggests a hereditary contribution, common exposures or shared risk factors among family members, or a combination of both.[2] Pathogenic variants in high-penetrance genes have been identified as the cause of inherited cancer risk in some colon cancer–prone families; these are estimated to account for only 5% to 6% of CRC cases overall.[3,4]
In addition, pathogenic variants in lower-penetrance genes may contribute to familial colon cancer risk. In such cases, gene-gene and gene-environment interactions may contribute to the development of CRC.
(Refer to the PDQ summaries on Colorectal Cancer Screening; Colorectal Cancer Prevention; Colon Cancer Treatment; and Rectal Cancer Treatment for more information about sporadic CRC.)
Colorectal Polyps as Precursors to Colorectal Cancer (CRC)
Colorectal tumors present with a broad spectrum of neoplasms, ranging from benign growths to invasive cancer, and are predominantly epithelial-derived tumors (i.e., adenomas or adenocarcinomas).
Transformation of any polyp into cancer goes through the adenoma-carcinoma sequence. Polyps that have traditionally been considered nonneoplastic include those of the hyperplastic, juvenile, hamartomatous, inflammatory, and lymphoid types. However, in certain circumstances, hamartomatous and juvenile polyps can progress into cancer.
Research, however, does suggest a substantial risk of colon cancer in individuals with juvenile polyposis syndrome and Peutz-Jeghers syndrome, although the nonadenomatous polyps associated with these syndromes have historically been viewed as nonneoplastic.[5-7]
Epidemiological studies have shown that a personal history of colon adenomas places one at an increased risk of developing colon cancer.[8]
Two complementary interpretations of this observation are as follows:
- The adenoma may reflect an innate or acquired tendency of the colon to form tumors.
- Adenomas are the primary precursor lesion of colon cancer.
More than 95% of CRCs are carcinomas, and about 95% of these are adenocarcinomas. It is well recognized that adenomatous polyps are benign tumors that may undergo malignant transformation. They have been classified into three histological types, with increasing malignant potential: tubular, tubulovillous, and villous. Adenocarcinomas are generally considered to arise from adenomas,[9-13] based upon the following important observations:
The following three characteristics of adenomas are highly correlated with the potential to transform into cancer:[14]
- Larger size.
- Villous pathology.
- The degree of dysplasia within the adenoma.
In addition, removal of adenomatous polyps is associated with reduced CRC incidence.[16,17] While most adenomas are polypoid, flat and depressed lesions may be more prevalent than previously recognized. Large, flat, and depressed lesions may be more likely to be severely dysplastic, although this remains to be clearly proven.[18,19] Specialized techniques may be needed to identify, biopsy, and remove such lesions.[20]
Family History as a Risk Factor for CRC
Some of the earliest studies of family history of CRC were those of Utah families that reported a higher percentage of deaths from CRC (3.9%) among the first-degree relatives (FDRs) of patients who had died from CRC than among sex-matched and age-matched controls (1.2%).[21] This difference has since been replicated in numerous studies that have consistently found that FDRs of affected cases are themselves at a twofold to threefold increased risk of CRC. Despite the various study designs (case-control, cohort), sampling frames, sample sizes, methods of data verification, analytic methods, and countries where the studies originated, the magnitude of risk is consistent.[22-27]
A systematic review and meta-analysis of familial CRC risk has been reported.[28] Of 24 studies included in the analysis, all but one reported an increased risk of CRC if there was an affected FDR. The relative risk (RR) for CRC in the pooled study was 2.25 (95% confidence interval [CI], 2.00–2.53) if there was an affected FDR. In 8 of 11 studies, if the index cancer arose in the colon, the risk was slightly higher than if it arose in the rectum. The pooled analysis revealed an RR in relatives of colon and rectal cancer patients of 2.42 (95% CI, 2.20–2.65) and 1.89 (95% CI, 1.62–2.21), respectively. The analysis did not reveal a difference in RR for colon cancer based on location of the tumor (right side vs. left side).
The number of affected family members and age at cancer diagnosis correlated with the CRC risk. In studies reporting more than one FDR with CRC, the RR was 3.76 (95% CI, 2.56–5.51). The highest RR was observed when the index case was diagnosed in individuals younger than 45 years (RR, 3.87; 95% CI, 2.40–6.22) compared with family members of index cases diagnosed at ages 45 to 59 years (RR, 2.25; 95% CI, 1.85–2.72), and to family members of index cases diagnosed at age 60 years or older (RR, 1.82; 95% CI, 1.47–2.25). In this meta-analysis, the familial risk of CRC associated with adenoma in an FDR was analyzed. The pooled analysis demonstrated an RR for CRC of 1.99 (95% CI, 1.55–2.55) in individuals who had an FDR with an adenoma.[28] This finding has been corroborated.[29] Other studies have reported that age at diagnosis of the adenoma influences the CRC risk, with younger age at adenoma diagnosis associated with higher RR.[30,31] As with any meta-analysis, there could be potential biases that might affect the results of the analysis, including incomplete and nonrandom ascertainment of studies included; publication bias; and heterogeneity between studies relative to design, target populations, and control selection. This study is reinforcement that there are significant associations between familial CRC risk, age at diagnosis of both CRC and adenomas, and multiplicity of affected family members.
| Family History | Relative Risk of CRC [28] | Absolute Risk (%) of CRC by Age 79 ya |
|---|---|---|
| CI = confidence interval; FDR = first-degree relative. | ||
| aData from the Surveillance, Epidemiology, and End Results Program database. | ||
| bThe absolute risks of CRC for individuals with affected relatives was calculated using the relative risks for CRC [28] and the absolute risk of CRC by age 79 yearsa. | ||
| No family history | 1 | 4a |
| One FDR with CRC | 2.3 (95% CI, 2.0–2.5) | 9b |
| More than one FDR with CRC | 4.3 (95% CI, 3.0–6.1) | 16b |
| One affected FDR diagnosed with CRC before age 45 y | 3.9 (95% CI, 2.4–6.2) | 15b |
| One FDR with colorectal adenoma | 2.0 (95% CI, 1.6–2.6) | 8b |
When the family history includes two or more relatives with CRC, the possibility of a genetic syndrome is increased substantially. The first step in this evaluation is a detailed review of the family history to determine the number of relatives affected, their relationship to each other, the age at which the CRC was diagnosed, the presence of multiple primary CRCs, and the presence of any other cancers (e.g., endometrial) consistent with an inherited CRC syndrome. (Refer to the Major Genetic Syndromes section of this summary for more information.) Computer models are now available to estimate the probability of developing CRC.[32] These models can be helpful in providing genetic counseling to individuals at average risk and high risk of developing cancer. In addition, at least three validated models are also available for predicting the probability of carrying a pathogenic variant in a mismatch repair (MMR) gene.[33-35]
Figure 1 shows the proportion of CRC cases that arise in various family risk settings.[36]
Inheritance of CRC Predisposition
Several genes associated with CRC risk have been identified; these are described in detail in the Colon Cancer Genes section of this summary. Almost all pathogenic variants known to cause a predisposition to CRC are inherited in an autosomal dominant fashion.[37] One example of autosomal recessive inheritance, MUTYH-associated polyposis (MAP), has been identified. (Refer to the MUTYH-Associated Polyposis [MAP] section of this summary for more information.) Thus, the family characteristics that suggest autosomal dominant inheritance of cancer predisposition are important indicators of high risk and of the possible presence of a cancer-predisposing pathogenic variant. These include the following:
- Vertical transmission of cancer predisposition in autosomal dominant conditions. (Vertical transmission refers to the presence of a genetic predisposition in sequential generations.)
- Inheritance risk of 50% for both male and female children. When a parent carries an autosomal dominant genetic predisposition, each child has a 50% chance of inheriting the predisposition. The risk is the same for both male and female children.
- Other clinical characteristics also suggest the presence of a hereditary CRC syndrome:
- Cancers in people with a hereditary predisposition typically occur at an earlier age than in sporadic cases.[38]
- A predisposition to CRC may include a predisposition to other cancers, such as endometrial cancer, as detailed in the Major Genetic Syndromes section of this summary.
- In addition, two or more primary cancers may occur in a single individual. These could be multiple primary cancers of the same type (e.g., two separate primary CRCs) or primary cancer of different types (e.g., colorectal and endometrial cancer in the same individual).
- The presence of nonneoplastic extracolonic features may suggest a hereditary colon cancer predisposition syndrome (e.g., congenital hypertrophy of the retinal pigment epithelium and desmoid tumors in familial adenomatous polyposis [FAP]).
- An uncommon tumor (e.g., adrenocortical carcinoma, sebaceous adenoma or carcinoma, and trichilemmoma) may serve as a clue to the presence of a hereditary cancer syndrome.
- The presence of multiple polyps may suggest a hereditary colon cancer predisposition syndrome. As susceptibility to oligopolyposis (as few as 10–15 polyps) has become apparent, clinicians, and gastrointestinal endoscopists in particular, may consider multigene (panel) testing of an ever-expanding list of genes associated with CRC. (Refer to Table 2, Genes Associated with a High Susceptibility of Colorectal Cancer, for more information.) Because oligopolyposis also involves diverse pathology (including hamartomas, sessile serrated polyps, and sessile serrated adenomas), careful attention to polyp count and polyp histologies helps to determine whether genetic testing and/or further clinical evaluation is appropriate.
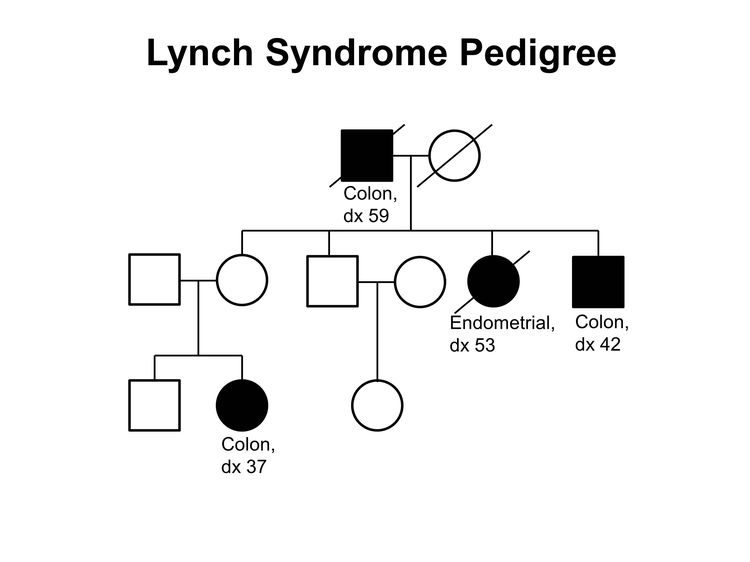
The two most common causes of hereditary CRC are FAP (including AFAP), due to germline pathogenic variants in the APC gene,[39-46] and Lynch syndrome (previously called hereditary nonpolyposis colorectal cancer [HNPCC]), which is caused by germline pathogenic variants in DNA MMR genes.[47-50] (Figure 2 depicts a classic family with Lynch syndrome, highlighting some of the indicators of hereditary CRC that are described above.) Many other families exhibit aggregation of CRC and/or adenomas, but with no apparent association with an identifiable hereditary syndrome, and are known collectively as familial CRC.[37]
Identification of Individuals at High Genetic Risk of CRC
National Comprehensive Cancer Network (NCCN) guidelines are updated annually to help identify patients who are appropriate for referral to cancer genetic counseling services. Furthermore, NCCN also provides cancer surveillance recommendations for hereditary cancer syndromes. The American College of Medical Genetics and Genomics and the National Society of Genetic Counselors have published a comprehensive set of personal/family history criteria to help identify at-risk individuals for referral to cancer genetics risk consultations.[51] These practice guidelines consider tumor types, other specific tumor features, and related criteria that would prompt a genetics referral. The authors state that these guidelines from ACMG/NSGC are intended to maximize referral of appropriate at-risk individuals to cancer genetic counseling services, but they are not meant to provide genetic testing or treatment recommendations. In addition, the authors note other sources that discuss updated/evolving genetic testing criteria for hereditary cancer syndromes (e.g., NCCN), and they acknowledge the increasing role of nongenetics professionals in facilitating genetic testing, especially to guide cancer treatment.[52]
When such persons are identified, options tailored to the patient situation are considered. (Refer to the Major Genetic Syndromes section of this summary for information on specific interventions for individual syndromes.)
At this time, the use of pathogenic variant testing to identify genetic susceptibility to CRC is not recommended as a screening measure in the general population. The rarity of pathogenic variants in CRC-associated genes and the limited sensitivity of current testing strategies render general population testing potentially misleading and not cost-effective.
Rather detailed recommendations for surveillance in FAP and Lynch syndrome have been provided by several organizations representing various medical specialties and societies. These organizations include the following:
- American Cancer Society.[53]
- United States Multisociety (American Gastroenterological Association and American Society for Gastrointestinal Endoscopy) Task Force on Colorectal Cancer.[54]
- American Society of Colon and Rectal Surgeons.[55]
- NCCN.[56]
- Gene Reviews.
- American College of Gastroenterology.[57]
- Society of Gynecologic Oncology and American College of Obstetrics & Gynecology.
The evidence bases for recommendations are generally included within the statements or guidelines. In many instances, these guidelines reflect expert opinion resting on studies that are rarely randomized prospective trials.
Early-onset CRC
The epidemiology of CRC with regard to age at diagnosis is shifting, with individuals increasingly being diagnosed before age 55 years,[1] often in the absence of polyposis and without a family history of CRC suggesting an inherited cancer syndrome.[58-60] (Refer to the PDQ summary on Colorectal Cancer Prevention for more information about CRC incidence trends in the general population.) One study that examined the prevalence of highly penetrant pathogenic variants in 450 individuals with early-onset CRC (mean age at diagnosis, 42.5 y) and a family history including at least one FDR with colon, endometrial, breast, ovarian, and/or pancreatic cancer identified 75 germline pathogenic or likely pathogenic variants in 72 patients (16%).[58] The spectrum of variants identified included Lynch syndrome and non-Lynch syndrome–associated genes, including several genes that have not traditionally been associated with CRC (e.g., BRCA1/BRCA2, ATM, CHEK2, PALB2, and CDKN2A). Given the high frequency and variety of hereditary cancer syndromes identified, the authors suggested that multigene (panel) testing in this population may be warranted.
In the absence of an additional family or personal history suggestive of Lynch syndrome, isolated cases of CRC diagnosed before age 36 years are uncommonly associated with MMR gene pathogenic variants. One study found MMR pathogenic variants in only 6.5% of such individuals,[59] whereas another study of patients with CRC younger than 50 years with no more than one FDR with CRC found abnormal microsatellite instability (MSI) in 21% of tumors and overrepresentation of defects in the PMS2 and MSH6 genes.[60] Therefore, isolated cases of very early-onset CRC in the absence of polyposis should be offered tumor screening with MSI/immunohistochemistry rather than proceeding directly to germline pathogenic variant analysis.
The use of polygenic risk scores (PRS) is being studied in the context of early-onset CRC in individuals who have tested negative for common CRC susceptibility variants (NCT02863107), with data from one large analysis [61] demonstrating that the predictive capacity of a 95-gene PRS may be particularly strong in assessing for CRC risk among young individuals (age, <50 y) who lack a family history of CRC in an FDR, and who would otherwise not be selected for early initiation of colonoscopic screening, by current practice.
Difficulties in Identifying a Family History of CRC Risk
The accuracy and completeness of family history data must be considered when using family history to assess individual risk in clinical practice and when identifying families appropriate for cancer research. A reported family history may be erroneous, or a person may be unaware of relatives with cancer.[62] Increased use of colonoscopy may result in fewer CRCs and more precancerous colon polyps in a family history. Individuals are much less likely to know about their family history of polyps (i.e., type of polyps and total number of polyps in their relatives) than they are to know about their family history of cancer. In addition, small family sizes and premature deaths may limit how informative a family history may be. Also, due to incomplete penetrance, some individuals may carry a genetic predisposition to CRC but do not develop cancer, giving the impression of skipped generations in a family tree.
Accuracy of patient-reported family history of colon cancer has been shown to be good, but it is not optimal. Patient report should be verified by obtaining medical records whenever possible, especially for reproductive tract cancers that may be relevant in identifying risk of Lynch syndrome and less reliably reported by some patients. (Refer to the Accuracy of the family history section in the PDQ summary on Cancer Genetics Risk Assessment and Counseling for more information.)
Several approaches are available to evaluate a patient with newly diagnosed CRC who may or may not be suspected of having a cancer genetics syndrome. The clinician may suspect a potential inherited disposition based on the family history and physical exam, and genetic tests are available to confirm these suspicions. The American College of Medical Genetics and Genomics has published guidelines for evaluating patients with suspected colon cancer susceptibility syndromes.[51] The guidelines aim to identify individuals whose clinical features warrant referral for genetics consultation. If an individual has multiple polyps (>20), depending on the histology, specific gene-directed testing can be a useful diagnostic tool. Similarly, if a patient’s clinical presentation is suspicious for Lynch syndrome, germline genetic testing can be directed towards this syndrome. However, diagnosis is more challenging when the clinical picture is less clear. Currently, tumor screening for Lynch syndrome is the most commonly accepted approach. However, increasingly, panels characterizing somatic variants in tumors are being utilized for a variety of clinical decisions.
A priori risk-assessment testing (which models risk based on a variety of factors, such as age at cancer onset and the spectrum of tumors in the family) may be an appropriate alternative in many cases. Application of such risk models does anticipate the use of multigene (panel) testing; however, their exact role remains to be established.
Molecular Events Associated With Colon Carcinogenesis
Much of our initial understanding of the molecular pathogenesis of CRC derived from rare hereditary CRC syndromes and revealed heterogeneity of CRC both molecularly and clinically. It is well accepted that most CRCs develop from adenomas. The transition from normal epithelium to adenoma to carcinoma is associated with acquired molecular events.[63-65] Presently, CRC can be separated into three categories based on similar molecular genetic features, suggesting divergent pathways of tumorigenesis: chromosomal instability (CIN), MSI, and CpG island methylator phenotype (CIMP). The understanding of the molecular genetic pathways of colorectal tumorigenesis is still evolving, and each new level of understanding has occurred in the context of the preceding level of knowledge. In addition, these pathways emerged from important clinical and histological heterogeneity of colorectal polyps and cancers. Thus, the introduction below captures the chronological evolution of our current understanding of colorectal tumorigenesis.
Chromosomal instability (CIN) pathway
Most CRCs develop through the CIN pathway. Key changes in CIN cancers include widespread alterations in chromosome number (aneuploidy) and frequent detectable losses at the molecular level of portions of chromosomes (loss of heterozygosity), such as 5q, 18q, and 17p; and pathogenic variants of the KRAS oncogene. The important genes involved in these chromosome losses are APC (5q), DCC/MADH2/MADH4 (18q), and TP53 (17p).[64,66] These chromosomal losses are indicative of genetic instability at the molecular and chromosomal levels.[65] Among the earliest and most common events in the colorectal tumor progression pathway is loss or pathogenic variant–inactivation of the APC gene. Pathogenic variant–inactivation of APC was first shown to be important to CRC in FAP, a hereditary CRC syndrome in which affected individuals harbor germline APC alterations, resulting in its loss of function and a dramatically increased incidence of colorectal polyps and cancers. Acquired or inherited pathogenic variants of DNA damage-repair genes, for example, base excision repair, nucleotide excision repair, double stranded repair, and MMR, also play a role in predisposing colorectal epithelial cells to pathogenic variants.
Microsatellite instability (MSI) pathway
Soon thereafter, a subset (10%–15%) of CRCs was identified that lacked evidence of chromosomal instability but exhibited aberrations in microsatellite repeat sequences,[67,68] a characteristic of tumors in patients with Lynch syndrome.[69] It was later found that hypermethylation of the MLH1 promoter is responsible for many sporadic CRCs with MSI. Germline variants in DNA MMR genes were discovered in patients with Lynch syndrome, whose CRCs frequently displayed MSI. Thus, the microsatellite instability pathway (MSI, sometimes referred to as MIN) was proposed.
The key characteristics of MSI cancers are that they have a largely intact chromosome complement and, as a result of defects in the DNA MMR system, more readily acquire pathogenic variants in important and often unique cancer-associated genes. These types of cancers are detectable at the molecular level by alterations in repeating units of DNA that occur normally throughout the genome, known as DNA microsatellites.
The rate of adenoma-to-carcinoma progression appears to be faster in microsatellite-unstable tumors than in microsatellite-stable tumors.[70] The foundation for this is the repeated reports of interval cancers in patients with recent, normal colonoscopy. Further support for this is seen in the serrated pathway (see below), in which high rates of interval cancer have also been observed.[71,72] Characteristic histological changes, such as increased mucin production, can be seen in tumors that demonstrate MSI, intratumoral T lymphocyte infiltration/Crohn-like reaction, etc., distinguishing the colorectal tumors in this pathway.
The knowledge derived from the study of inherited CRC syndromes has provided important clues regarding the molecular events that mediate tumor initiation and tumor progression in people without germline abnormalities. Among the earliest events in the colorectal tumor progression pathway (both MSI and CIN) is loss of function of the APC gene product.
CpG island methylator phenotype (CIMP) and the serrated polyposis pathway
Beginning in the 1980s, studies began reporting an increased risk of CRC in patients with hyperplastic polyposis syndrome (HPS), now referred to as serrated polyposis syndrome (SPS).[6,7,73-78] Only a minority of SPS appear to be familial, but no common germline variant has been identified in these families to date. A comparison of the hyperplastic polyps (HPs) found in SPS patients and controls revealed that SPS polyps are histologically distinct and are similar to previously described serrated adenomas, polyps with features of HPs and adenomatous polyps (APs).[79] This led to observations that these sessile serrated adenomas (SSA) tend to occur in the right colon, where they are frequently large and sessile, and exhibit increased proliferation, dilation and serration of the crypt bases, decreased endocrine cells, and lack of dysplasia.[80]
Further histological characterization of serrated polyps led to subtypes: traditional serrated adenomas (TSA), mixed serrated polyps (MP), and more recently, sessile serrated adenoma/sessile serrated polyp (SSA/SSP).[81] TSAs are characterized by a protuberant morphology, ectopic crypt formation (suggestive of deficient bone morphogenetic protein signaling), and villiform and dysplastic histopathology.[80,82] TSAs are not simply SSAs with dysplasia, and evidence that SSAs are precursors of TSAs is lacking. MPs have overlapping features of HPs, SSAs, and TSAs.
In colonoscopy screening studies, large serrated polyps were strongly and independently associated with the development of advanced colorectal neoplasms, while left-sided HPs were not. The term SSA has been a concern to clinicians as these characteristically lack nuclear atypia, the traditional hallmark of adenomas, but rather are termed adenomas due to other architectural features. The classification of SSA is supported by the knowledge that the molecular characteristics denote an increased cancer risk.[79,83,84]
While APs in Lynch syndrome patients can exhibit MSI, sporadic adenomas rarely do. However, serrated polyps with dysplasia can exhibit MSI with hypermethylation of the MLH1 promoter. Large (>1 cm) serrated polyps carry greater cancer risk than do conventional hyperplastic polyps and, when developing into cancers, characteristically exhibit MSI.[82,85-87] In a review of resected serrated polyps with a malignant focus, all of the polyps originated in the right colon and were SSAs.[85] The malignant foci were MSI and demonstrated loss of MLH1 immunoreactivity, suggesting an association between SSAs and sporadic MSI colon cancers.
The MSI seen in sporadic CRCs is due to hypermethylation of the promoter of MLH1, which abrogates its expression. As promoter regions of other tumor suppressor genes were “silenced” through hypermethylation, cancer genome studies of CRC ensued. These showed a consistent pattern of hypermethylation in the evaluated genes in approximately 50% of CRCs.[88] Studies of larger numbers of unselected CRC patients show that a minority of CRCs (20%–30%) demonstrate CIMP, defined as hypermethylation of two or more of the CpG islands in MINT1, MINT2, MINT31, CDKN2A (p16), and MLH1.[89,90] The term CIMP was coined to classify these cancers, which shared clinical features. Early attempts to differentiate CIMP-positive and CIMP-negative CRCs were unsuccessful.[91] However, subsequent studies using unbiased hierarchical cluster analysis of heavily methylated genes in CRCs and a population-based study design successfully identified unique clinical and molecular characteristics supporting a CIMP pathway.[88,92]
CIMP-high CRCs were much more likely (82.1%; P < .0001) to express MSI than were microsatellite-stable CRCs (24.4%; P < .0001).[88] In one study, microsatellite-stable, CIMP-high (>2 CIMP markers mentioned above) colorectal tumors were significantly more associated with BRAF V600E variants, KRAS2 variants, proximal site, higher American Joint Committee on Cancer stage, older patient age, poor differentiation, and mucinous histology than were CIMP-low (<2 CIMP markers mentioned above) colorectal tumors.[88] Microsatellite-unstable, CIMP-high colorectal tumors were significantly more associated with BRAF V600E pathogenic variants, proximal site, older patient age, and absence of KRAS2 pathogenic variants than were microsatellite unstable, CIMP-low tumors.[88] There was a significantly greater presence of BRAF V600E pathogenic variants in CIMP-high colorectal tumors regardless of MSI.[88] Thus, unlike a previous study that questioned the biological significance of CIMP once unstable colorectal tumors were excluded,[91] this study demonstrated several clinicopathologic variables were indeed associated with CIMP in microsatellite-stable and microsatellite-unstable colorectal tumors.[88]
Studies of polyps revealed CIMP-positive polyps in SPS patients and most frequently in right-sided SSAs.[72,93-96] More recently, a hotspot BRAF pathogenic variant (V600E) was found to be common in MSI colon cancers and serrated polyps.[97-99] A BRAF pathogenic variant is absent in CRCs from Lynch syndrome patients and is rare in sporadic adenomatous colorectal polyps, but it is present in the vast majority of serrated polyps, especially SSAs.[94,96,100-102] CIMP positivity is commonly found in microvesicular hyperplastic polyps (MVHP), suggesting progression of MVHP to SSA and then to colon cancer.[94]
Conclusion
The characterization of CIMP CRCs and evidence that MSI occurs later in the adenoma-carcinoma sequence leads to modification of the previous colorectal tumorigenesis model, which was comprised of two pathways: MSI (MIN) and CIN. There is much overlap between the MSI and CIMP pathways. At the heart of the CIMP pathway are serrated polyps harboring BRAF pathogenic variants. The CIN pathway is characterized by AP precursors of which the vast majority harbor APC pathogenic variants that occur early in the pathway.
References
- American Cancer Society: Cancer Facts and Figures 2024. American Cancer Society, 2024. Available online. Last accessed December 30, 2024.
- Kanth P, Grimmett J, Champine M, et al.: Hereditary Colorectal Polyposis and Cancer Syndromes: A Primer on Diagnosis and Management. Am J Gastroenterol 112 (10): 1509-1525, 2017. [PUBMED Abstract]
- Lynch HT, Smyrk TC, Watson P, et al.: Genetics, natural history, tumor spectrum, and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology 104 (5): 1535-49, 1993. [PUBMED Abstract]
- Rustgi AK: The genetics of hereditary colon cancer. Genes Dev 21 (20): 2525-38, 2007. [PUBMED Abstract]
- Howe JR, Mitros FA, Summers RW: The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol 5 (8): 751-6, 1998. [PUBMED Abstract]
- Jeevaratnam P, Cottier DS, Browett PJ, et al.: Familial giant hyperplastic polyposis predisposing to colorectal cancer: a new hereditary bowel cancer syndrome. J Pathol 179 (1): 20-5, 1996. [PUBMED Abstract]
- Rashid A, Houlihan PS, Booker S, et al.: Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology 119 (2): 323-32, 2000. [PUBMED Abstract]
- Neugut AI, Jacobson JS, DeVivo I: Epidemiology of colorectal adenomatous polyps. Cancer Epidemiol Biomarkers Prev 2 (2): 159-76, 1993 Mar-Apr. [PUBMED Abstract]
- Shinya H, Wolff WI: Morphology, anatomic distribution and cancer potential of colonic polyps. Ann Surg 190 (6): 679-83, 1979. [PUBMED Abstract]
- Fenoglio CM, Lane N: The anatomical precursor of colorectal carcinoma. Cancer 34 (3): suppl:819-23, 1974. [PUBMED Abstract]
- Morson B: President's address. The polyp-cancer sequence in the large bowel. Proc R Soc Med 67 (6): 451-7, 1974. [PUBMED Abstract]
- Muto T, Bussey HJ, Morson BC: The evolution of cancer of the colon and rectum. Cancer 36 (6): 2251-70, 1975. [PUBMED Abstract]
- Stryker SJ, Wolff BG, Culp CE, et al.: Natural history of untreated colonic polyps. Gastroenterology 93 (5): 1009-13, 1987. [PUBMED Abstract]
- O'Brien MJ, Winawer SJ, Zauber AG, et al.: The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology 98 (2): 371-9, 1990. [PUBMED Abstract]
- Winawer SJ, Stewart ET, Zauber AG, et al.: A comparison of colonoscopy and double-contrast barium enema for surveillance after polypectomy. National Polyp Study Work Group. N Engl J Med 342 (24): 1766-72, 2000. [PUBMED Abstract]
- Winawer SJ, Zauber AG, Ho MN, et al.: Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med 329 (27): 1977-81, 1993. [PUBMED Abstract]
- Müller AD, Sonnenberg A: Prevention of colorectal cancer by flexible endoscopy and polypectomy. A case-control study of 32,702 veterans. Ann Intern Med 123 (12): 904-10, 1995. [PUBMED Abstract]
- O'brien MJ, Winawer SJ, Zauber AG, et al.: Flat adenomas in the National Polyp Study: is there increased risk for high-grade dysplasia initially or during surveillance? Clin Gastroenterol Hepatol 2 (10): 905-11, 2004. [PUBMED Abstract]
- Zauber AG, O'Brien MJ, Winawer SJ: On finding flat adenomas: is the search worth the gain? Gastroenterology 122 (3): 839-40, 2002. [PUBMED Abstract]
- Rembacken BJ, Fujii T, Cairns A, et al.: Flat and depressed colonic neoplasms: a prospective study of 1000 colonoscopies in the UK. Lancet 355 (9211): 1211-4, 2000. [PUBMED Abstract]
- Woolf CM: A genetic study of carcinoma of the large intestine. Am J Hum Genet 10 (1): 42-7, 1958. [PUBMED Abstract]
- Fuchs CS, Giovannucci EL, Colditz GA, et al.: A prospective study of family history and the risk of colorectal cancer. N Engl J Med 331 (25): 1669-74, 1994. [PUBMED Abstract]
- Slattery ML, Kerber RA: Family history of cancer and colon cancer risk: the Utah Population Database. J Natl Cancer Inst 86 (21): 1618-26, 1994. [PUBMED Abstract]
- Negri E, Braga C, La Vecchia C, et al.: Family history of cancer and risk of colorectal cancer in Italy. Br J Cancer 77 (1): 174-9, 1998. [PUBMED Abstract]
- St John DJ, McDermott FT, Hopper JL, et al.: Cancer risk in relatives of patients with common colorectal cancer. Ann Intern Med 118 (10): 785-90, 1993. [PUBMED Abstract]
- Duncan JL, Kyle J: Family incidence of carcinoma of the colon and rectum in north-east Scotland. Gut 23 (2): 169-71, 1982. [PUBMED Abstract]
- Rozen P, Fireman Z, Figer A, et al.: Family history of colorectal cancer as a marker of potential malignancy within a screening program. Cancer 60 (2): 248-54, 1987. [PUBMED Abstract]
- Johns LE, Houlston RS: A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol 96 (10): 2992-3003, 2001. [PUBMED Abstract]
- Cottet V, Pariente A, Nalet B, et al.: Colonoscopic screening of first-degree relatives of patients with large adenomas: increased risk of colorectal tumors. Gastroenterology 133 (4): 1086-92, 2007. [PUBMED Abstract]
- Winawer SJ, Zauber AG, Gerdes H, et al.: Risk of colorectal cancer in the families of patients with adenomatous polyps. National Polyp Study Workgroup. N Engl J Med 334 (2): 82-7, 1996. [PUBMED Abstract]
- Ahsan H, Neugut AI, Garbowski GC, et al.: Family history of colorectal adenomatous polyps and increased risk for colorectal cancer. Ann Intern Med 128 (11): 900-5, 1998. [PUBMED Abstract]
- Win AK, Macinnis RJ, Hopper JL, et al.: Risk prediction models for colorectal cancer: a review. Cancer Epidemiol Biomarkers Prev 21 (3): 398-410, 2012. [PUBMED Abstract]
- Chen S, Wang W, Lee S, et al.: Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 296 (12): 1479-87, 2006. [PUBMED Abstract]
- Balmaña J, Stockwell DH, Steyerberg EW, et al.: Prediction of MLH1 and MSH2 mutations in Lynch syndrome. JAMA 296 (12): 1469-78, 2006. [PUBMED Abstract]
- Barnetson RA, Tenesa A, Farrington SM, et al.: Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med 354 (26): 2751-63, 2006. [PUBMED Abstract]
- Burt RW: Colon cancer screening. Gastroenterology 119 (3): 837-53, 2000. [PUBMED Abstract]
- Burt RW, Petersen GM: Familial colorectal cancer: diagnosis and management. In: Young GP, Rozen P, Levin B, eds.: Prevention and Early Detection of Colorectal Cancer. WB Saunders, 1996, pp 171-194.
- Mork ME, You YN, Ying J, et al.: High Prevalence of Hereditary Cancer Syndromes in Adolescents and Young Adults With Colorectal Cancer. J Clin Oncol 33 (31): 3544-9, 2015. [PUBMED Abstract]
- Kinzler KW, Nilbert MC, Su LK, et al.: Identification of FAP locus genes from chromosome 5q21. Science 253 (5020): 661-5, 1991. [PUBMED Abstract]
- Groden J, Thliveris A, Samowitz W, et al.: Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66 (3): 589-600, 1991. [PUBMED Abstract]
- Leppert M, Burt R, Hughes JP, et al.: Genetic analysis of an inherited predisposition to colon cancer in a family with a variable number of adenomatous polyps. N Engl J Med 322 (13): 904-8, 1990. [PUBMED Abstract]
- Spirio L, Olschwang S, Groden J, et al.: Alleles of the APC gene: an attenuated form of familial polyposis. Cell 75 (5): 951-7, 1993. [PUBMED Abstract]
- Brensinger JD, Laken SJ, Luce MC, et al.: Variable phenotype of familial adenomatous polyposis in pedigrees with 3' mutation in the APC gene. Gut 43 (4): 548-52, 1998. [PUBMED Abstract]
- Soravia C, Berk T, Madlensky L, et al.: Genotype-phenotype correlations in attenuated adenomatous polyposis coli. Am J Hum Genet 62 (6): 1290-301, 1998. [PUBMED Abstract]
- Pedemonte S, Sciallero S, Gismondi V, et al.: Novel germline APC variants in patients with multiple adenomas. Genes Chromosomes Cancer 22 (4): 257-67, 1998. [PUBMED Abstract]
- Sieber OM, Lamlum H, Crabtree MD, et al.: Whole-gene APC deletions cause classical familial adenomatous polyposis, but not attenuated polyposis or "multiple" colorectal adenomas. Proc Natl Acad Sci U S A 99 (5): 2954-8, 2002. [PUBMED Abstract]
- Leach FS, Nicolaides NC, Papadopoulos N, et al.: Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75 (6): 1215-25, 1993. [PUBMED Abstract]
- Papadopoulos N, Nicolaides NC, Wei YF, et al.: Mutation of a mutL homolog in hereditary colon cancer. Science 263 (5153): 1625-9, 1994. [PUBMED Abstract]
- Nicolaides NC, Papadopoulos N, Liu B, et al.: Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371 (6492): 75-80, 1994. [PUBMED Abstract]
- Miyaki M, Konishi M, Tanaka K, et al.: Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 17 (3): 271-2, 1997. [PUBMED Abstract]
- Hampel H, Bennett RL, Buchanan A, et al.: A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med 17 (1): 70-87, 2015. [PUBMED Abstract]
- Bashford MT, Kohlman W, Everett J, et al.: Addendum: A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med 21 (12): 2844, 2019. [PUBMED Abstract]
- Smith RA, Cokkinides V, Eyre HJ: American Cancer Society guidelines for the early detection of cancer, 2006. CA Cancer J Clin 56 (1): 11-25; quiz 49-50, 2006 Jan-Feb. [PUBMED Abstract]
- Winawer S, Fletcher R, Rex D, et al.: Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology 124 (2): 544-60, 2003. [PUBMED Abstract]
- Church J, Simmang C; Standards Task Force, et al.: Practice parameters for the treatment of patients with dominantly inherited colorectal cancer (familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer). Dis Colon Rectum 46 (8): 1001-12, 2003. [PUBMED Abstract]
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal, Endometrial, and Gastric. Version 3.2024. Plymouth Meeting, PA: National Comprehensive Cancer Network, 2024. Available with free registration. Last accessed December 13, 2024.
- Syngal S, Brand RE, Church JM, et al.: ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 110 (2): 223-62; quiz 263, 2015. [PUBMED Abstract]
- Pearlman R, Frankel WL, Swanson B, et al.: Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol 3 (4): 464-471, 2017. [PUBMED Abstract]
- Jasperson KW, Vu TM, Schwab AL, et al.: Evaluating Lynch syndrome in very early onset colorectal cancer probands without apparent polyposis. Fam Cancer 9 (2): 99-107, 2010. [PUBMED Abstract]
- Goel A, Nagasaka T, Spiegel J, et al.: Low frequency of Lynch syndrome among young patients with non-familial colorectal cancer. Clin Gastroenterol Hepatol 8 (11): 966-71, 2010. [PUBMED Abstract]
- Archambault AN, Su YR, Jeon J, et al.: Cumulative Burden of Colorectal Cancer-Associated Genetic Variants Is More Strongly Associated With Early-Onset vs Late-Onset Cancer. Gastroenterology 158 (5): 1274-1286.e12, 2020. [PUBMED Abstract]
- Glanz K, Grove J, Le Marchand L, et al.: Underreporting of family history of colon cancer: correlates and implications. Cancer Epidemiol Biomarkers Prev 8 (7): 635-9, 1999. [PUBMED Abstract]
- Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell 61 (5): 759-67, 1990. [PUBMED Abstract]
- Vogelstein B, Kinzler KW: The multistep nature of cancer. Trends Genet 9 (4): 138-41, 1993. [PUBMED Abstract]
- Lengauer C, Kinzler KW, Vogelstein B: Genetic instabilities in human cancers. Nature 396 (6712): 643-9, 1998. [PUBMED Abstract]
- Kinzler KW, Vogelstein B: Colorectal tumors. In: Vogelstein B, Kinzler KW, eds.: The Genetic Basis of Human Cancer. 2nd ed. McGraw-Hill, 2002, pp 583-612.
- Thibodeau SN, Bren G, Schaid D: Microsatellite instability in cancer of the proximal colon. Science 260 (5109): 816-9, 1993. [PUBMED Abstract]
- Ionov Y, Peinado MA, Malkhosyan S, et al.: Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363 (6429): 558-61, 1993. [PUBMED Abstract]
- Peltomäki P, Lothe RA, Aaltonen LA, et al.: Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res 53 (24): 5853-5, 1993. [PUBMED Abstract]
- Jass JR, Cottier DS, Pokos V, et al.: Mixed epithelial polyps in association with hereditary non-polyposis colorectal cancer providing an alternative pathway of cancer histogenesis. Pathology 29 (1): 28-33, 1997. [PUBMED Abstract]
- Jass JR: Serrated route to colorectal cancer: back street or super highway? J Pathol 193 (3): 283-5, 2001. [PUBMED Abstract]
- Wynter CV, Walsh MD, Higuchi T, et al.: Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut 53 (4): 573-80, 2004. [PUBMED Abstract]
- Bengoechea O, Martínez-Peñuela JM, Larrínaga B, et al.: Hyperplastic polyposis of the colorectum and adenocarcinoma in a 24-year-old man. Am J Surg Pathol 11 (4): 323-7, 1987. [PUBMED Abstract]
- Hyman NH, Anderson P, Blasyk H: Hyperplastic polyposis and the risk of colorectal cancer. Dis Colon Rectum 47 (12): 2101-4, 2004. [PUBMED Abstract]
- Leggett BA, Devereaux B, Biden K, et al.: Hyperplastic polyposis: association with colorectal cancer. Am J Surg Pathol 25 (2): 177-84, 2001. [PUBMED Abstract]
- McCann BG: A case of metaplastic polyposis of the colon associated with focal adenomatous change and metachronous adenocarcinomas. Histopathology 13 (6): 700-2, 1988. [PUBMED Abstract]
- Place RJ, Simmang CL: Hyperplastic-adenomatous polyposis syndrome. J Am Coll Surg 188 (5): 503-7, 1999. [PUBMED Abstract]
- Koide N, Saito Y, Fujii T, et al.: A case of hyperplastic polyposis of the colon with adenocarcinomas in hyperplastic polyps after long-term follow-up. Endoscopy 34 (6): 499-502, 2002. [PUBMED Abstract]
- Torlakovic E, Snover DC: Serrated adenomatous polyposis in humans. Gastroenterology 110 (3): 748-55, 1996. [PUBMED Abstract]
- Torlakovic EE, Gomez JD, Driman DK, et al.: Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA). Am J Surg Pathol 32 (1): 21-9, 2008. [PUBMED Abstract]
- Snover DC, Jass JR, Fenoglio-Preiser C, et al.: Serrated polyps of the large intestine: a morphologic and molecular review of an evolving concept. Am J Clin Pathol 124 (3): 380-91, 2005. [PUBMED Abstract]
- Lash RH, Genta RM, Schuler CM: Sessile serrated adenomas: prevalence of dysplasia and carcinoma in 2139 patients. J Clin Pathol 63 (8): 681-6, 2010. [PUBMED Abstract]
- Torlakovic E, Skovlund E, Snover DC, et al.: Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol 27 (1): 65-81, 2003. [PUBMED Abstract]
- Jass JR, Baker K, Zlobec I, et al.: Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a 'fusion' pathway to colorectal cancer. Histopathology 49 (2): 121-31, 2006. [PUBMED Abstract]
- Goldstein NS: Small colonic microsatellite unstable adenocarcinomas and high-grade epithelial dysplasias in sessile serrated adenoma polypectomy specimens: a study of eight cases. Am J Clin Pathol 125 (1): 132-45, 2006. [PUBMED Abstract]
- Lu FI, van Niekerk de W, Owen D, et al.: Longitudinal outcome study of sessile serrated adenomas of the colorectum: an increased risk for subsequent right-sided colorectal carcinoma. Am J Surg Pathol 34 (7): 927-34, 2010. [PUBMED Abstract]
- Schreiner MA, Weiss DG, Lieberman DA: Proximal and large hyperplastic and nondysplastic serrated polyps detected by colonoscopy are associated with neoplasia. Gastroenterology 139 (5): 1497-502, 2010. [PUBMED Abstract]
- Toyota M, Ahuja N, Ohe-Toyota M, et al.: CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 96 (15): 8681-6, 1999. [PUBMED Abstract]
- Ahuja N, Mohan AL, Li Q, et al.: Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res 57 (16): 3370-4, 1997. [PUBMED Abstract]
- Samowitz WS, Albertsen H, Herrick J, et al.: Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 129 (3): 837-45, 2005. [PUBMED Abstract]
- Yamashita K, Dai T, Dai Y, et al.: Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell 4 (2): 121-31, 2003. [PUBMED Abstract]
- Weisenberger DJ, Siegmund KD, Campan M, et al.: CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38 (7): 787-93, 2006. [PUBMED Abstract]
- Chan AO, Issa JP, Morris JS, et al.: Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol 160 (2): 529-36, 2002. [PUBMED Abstract]
- Kambara T, Simms LA, Whitehall VL, et al.: BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 53 (8): 1137-44, 2004. [PUBMED Abstract]
- O'Brien MJ, Yang S, Clebanoff JL, et al.: Hyperplastic (serrated) polyps of the colorectum: relationship of CpG island methylator phenotype and K-ras mutation to location and histologic subtype. Am J Surg Pathol 28 (4): 423-34, 2004. [PUBMED Abstract]
- Yang S, Farraye FA, Mack C, et al.: BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol 28 (11): 1452-9, 2004. [PUBMED Abstract]
- Chan TL, Zhao W, Leung SY, et al.: BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 63 (16): 4878-81, 2003. [PUBMED Abstract]
- Rajagopalan H, Bardelli A, Lengauer C, et al.: Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418 (6901): 934, 2002. [PUBMED Abstract]
- Yuen ST, Davies H, Chan TL, et al.: Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 62 (22): 6451-5, 2002. [PUBMED Abstract]
- Deng G, Bell I, Crawley S, et al.: BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res 10 (1 Pt 1): 191-5, 2004. [PUBMED Abstract]
- McGivern A, Wynter CV, Whitehall VL, et al.: Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer 3 (2): 101-7, 2004. [PUBMED Abstract]
- Wang L, Cunningham JM, Winters JL, et al.: BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 63 (17): 5209-12, 2003. [PUBMED Abstract]
Colorectal Cancer Susceptibility Genes
Major Genes
Major genes are defined as those that are necessary and sufficient for disease causation, with important pathogenic variants (e.g., nonsense, missense, frameshift) of the gene as causal mechanisms. Major genes are typically considered those that are involved in single-gene disorders, and the diseases caused by major genes are often relatively rare. Most pathogenic variants in major genes lead to a very high risk of disease, and environmental contributions are often difficult to recognize.[1] Historically, most major colon cancer susceptibility genes have been identified by linkage analysis using high-risk families; thus, these criteria were fulfilled by definition, as a consequence of the study design.
The functions of the major colorectal (CRC) cancer genes have been reasonably well characterized over the past decade.[2] Tumor suppressor genes constitute the most important class of genes responsible for hereditary cancer syndromes and represent the class of genes responsible for familial adenomatous polyposis (FAP), Lynch syndrome, and juvenile polyposis syndrome (JPS), among others. Table 2 summarizes the genes that confer a substantial risk of CRC, with their corresponding diseases.
| Gene | Syndrome | Hereditary Pattern | Predominant Cancers |
|---|---|---|---|
| FAP = familial adenomatous polyposis; JPS = juvenile polyposis syndrome; PJS = Peutz-Jeghers syndrome; PPAP = polymerase proofreading–associated polyposis. | |||
| APC | FAP, AFAP | Dominant | Colorectal, small bowel, gastric, etc. |
| TP53 (p53) | Li-Fraumeni | Dominant | Multiple (including colorectal) |
| STK11 (LKB1) | PJS | Dominant | Multiple (including colorectal, small bowel, pancreas) |
| PTEN | Cowden | Dominant | Multiple (including colorectal) |
| BMPR1A, SMAD4 (MADH/DPC4) | JPS | Dominant | Gastric and colorectal |
| MLH1, MSH2, MSH6, PMS2, EPCAM | Lynch syndrome | Dominant | Multiple (including colorectal, endometrial, and others) |
| MUTYH (MYH) | MUTYH-associated polyposis | Recessive | Colorectal |
| POLD1, POLE | PPAP | Dominant | Colorectal, endometrial |
De Novo Pathogenic Variant Rate
Until the 1990s, the diagnosis of genetically inherited polyposis syndromes was based on clinical manifestations and family history. Now that some of the genes involved in these syndromes have been identified, a few studies have attempted to estimate the spontaneous pathogenic variant rate (de novo pathogenic variant rate) in these populations. Interestingly, FAP, JPS, Peutz-Jeghers syndrome, Cowden syndrome, and Bannayan-Riley-Ruvalcaba syndrome are all thought to have high rates of spontaneous pathogenic variants, in the 25% to 30% range,[3-5] while estimates of de novo pathogenic variants in the MMR genes associated with Lynch syndrome are thought to be low, in the 0.9% to 5% range.[6-8] These estimates of spontaneous pathogenic variant rates in Lynch syndrome seem to overlap with the estimates of nonpaternity rates in various populations (0.6% to 3.3%),[9-11] making the de novo pathogenic variant rate for Lynch syndrome seem quite low in contrast to the relatively high rates in the other polyposis syndromes.
Genetic Polymorphisms and CRC Risk
It is widely acknowledged that the familial clustering of colon cancer also occurs outside of the setting of well-characterized colon cancer family syndromes.[12] Based on epidemiological studies, the risk of colon cancer in a first-degree relative (FDR) of an affected individual can increase an individual’s lifetime risk of colon cancer 2-fold to 4.3-fold.[13] The relative risk (RR) and absolute risk of CRC for different family history categories is estimated in Table 1. In addition, the lifetime risk of colon cancer also increases in FDRs of individuals with colon adenomas.[14] The magnitude of risk depends on the age at diagnosis of the index case, the degree of relatedness of the index case to the at-risk case, and the number of affected relatives. It is currently believed that many of the moderate- and low-risk cases are influenced by alterations in single low-penetrance genes or combinations of low-penetrance genes.[15] Given the public health impact of identifying the etiology of this increased risk, an intense search for the responsible genes is under way.
Each locus would be expected to have a relatively small effect on CRC risk and would not produce the dramatic familial aggregation seen in Lynch syndrome or FAP. However, in combination with other common genetic loci and/or environmental factors, variants of this kind might significantly alter CRC risk. These types of genetic variations are often referred to as polymorphisms. Most loci that are polymorphic have no influence on disease risk or human traits (benign polymorphisms), while those that are associated with a difference in risk of disease or a human trait (however subtle) are sometimes termed disease-associated polymorphisms or functionally relevant polymorphisms. When such variation involves changes in single nucleotides of DNA they are referred to as single nucleotide variants (SNVs).
Several genome-wide association studies (GWAS) have been conducted with relatively large, unselected series of patients with CRC, who have been evaluated for patterns of polymorphisms in candidate and anonymous genes throughout the genome.[16-19] The use of genome-wide scans in thousands of CRC cases and controls has led to the discovery of multiple common, low-risk CRC SNVs, which can be found in the National Human Genome Research Institute GWAS catalog. For more information on GWAS, see Cancer Genetics Overview.
A goal of these studies has been to identify SNVs that may confer mildly increased CRC risks. This risk stratification could potentially enhance usability of CRC screening by influencing patient decision making regarding the ages to begin/end CRC screening and the amount of time between screening studies. There is increasing interest about using SNVs to expand germline risk assessment. This could be expanded from only searching for monogenic forms of CRC predisposition with high-/moderate-penetrance to searching for forms of CRC predisposition with polygenic risk, which may have broader applicability in the general population. Additionally, multiple studies have examined if polygenic risk scores (PRS) can be used to personalize CRC risk assessment in individuals with average risk of developing CRC.[20,21] While the growing body of data on CRC risk SNVs and PRS have been promising, PRS are not currently used in routine clinical settings and are not considered clinically actionable to guide the use of CRC screening procedures. Formal implementation studies are warranted to analyze how PRS can guide CRC risk assessment and screening in routine clinical care.
APC I1307K
The APC I1307K polymorphism deserves special mention, given that it is commonly identified in individuals of Ashkenazi Jewish ancestry undergoing multigene (panel) testing [22,23] and is associated with an increased risk of CRC; however, it does not cause colonic polyposis. The I1307K polymorphism occurs almost exclusively in people of Ashkenazi Jewish descent and results in a twofold increased risk of colonic adenomas and adenocarcinomas compared with the general population.[24,25] The I1307K polymorphism results from a transition from T to A at nucleotide 3920 in the APC gene and appears to create a region of hypermutability by virtue of the fact that this results in an A8 microsatellite coding sequence.[24] Although clinical assays to assess for the APC I1307K polymorphism are currently available, the associated CRC risk is not high enough to support their routine use. Based on currently available data, it is not yet known whether the I1307K carrier status should guide decisions regarding the age to initiate screening, the frequency of screening, or the choice of screening strategy.
References
- Caporaso N, Goldstein A: Cancer genes: single and susceptibility: exposing the difference. Pharmacogenetics 5 (2): 59-63, 1995. [PUBMED Abstract]
- Vogelstein B, Kinzler KW: Cancer genes and the pathways they control. Nat Med 10 (8): 789-99, 2004. [PUBMED Abstract]
- Aretz S, Uhlhaas S, Caspari R, et al.: Frequency and parental origin of de novo APC mutations in familial adenomatous polyposis. Eur J Hum Genet 12 (1): 52-8, 2004. [PUBMED Abstract]
- Westerman AM, Entius MM, Boor PP, et al.: Novel mutations in the LKB1/STK11 gene in Dutch Peutz-Jeghers families. Hum Mutat 13 (6): 476-81, 1999. [PUBMED Abstract]
- Schreibman IR, Baker M, Amos C, et al.: The hamartomatous polyposis syndromes: a clinical and molecular review. Am J Gastroenterol 100 (2): 476-90, 2005. [PUBMED Abstract]
- Morak M, Laner A, Scholz M, et al.: Report on de-novo mutation in the MSH2 gene as a rare event in hereditary nonpolyposis colorectal cancer. Eur J Gastroenterol Hepatol 20 (11): 1101-5, 2008. [PUBMED Abstract]
- Plasilova M, Zhang J, Okhowat R, et al.: A de novo MLH1 germ line mutation in a 31-year-old colorectal cancer patient. Genes Chromosomes Cancer 45 (12): 1106-10, 2006. [PUBMED Abstract]
- Win AK, Jenkins MA, Buchanan DD, et al.: Determining the frequency of de novo germline mutations in DNA mismatch repair genes. J Med Genet 48 (8): 530-4, 2011. [PUBMED Abstract]
- Anderson KG: How well does paternity confidence match actual paternity? Evidence from worldwide nonpaternity rates. Curr Anthropol 47 (3): 513-20, 2006. Also available online. Last accessed May 13, 2025.
- Sasse G, Müller H, Chakraborty R, et al.: Estimating the frequency of nonpaternity in Switzerland. Hum Hered 44 (6): 337-43, 1994 Nov-Dec. [PUBMED Abstract]
- Voracek M, Haubner T, Fisher ML: Recent decline in nonpaternity rates: a cross-temporal meta-analysis. Psychol Rep 103 (3): 799-811, 2008. [PUBMED Abstract]
- Burt RW, Bishop DT, Lynch HT, et al.: Risk and surveillance of individuals with heritable factors for colorectal cancer. WHO Collaborating Centre for the Prevention of Colorectal Cancer. Bull World Health Organ 68 (5): 655-65, 1990. [PUBMED Abstract]
- Butterworth AS, Higgins JP, Pharoah P: Relative and absolute risk of colorectal cancer for individuals with a family history: a meta-analysis. Eur J Cancer 42 (2): 216-27, 2006. [PUBMED Abstract]
- Johns LE, Houlston RS: A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol 96 (10): 2992-3003, 2001. [PUBMED Abstract]
- Wei C, Peng B, Han Y, et al.: Mutations of HNRNPA0 and WIF1 predispose members of a large family to multiple cancers. Fam Cancer 14 (2): 297-306, 2015. [PUBMED Abstract]
- Houlston RS, Webb E, Broderick P, et al.: Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 40 (12): 1426-35, 2008. [PUBMED Abstract]
- Houlston RS, Cheadle J, Dobbins SE, et al.: Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet 42 (11): 973-7, 2010. [PUBMED Abstract]
- Whiffin N, Hosking FJ, Farrington SM, et al.: Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet 23 (17): 4729-37, 2014. [PUBMED Abstract]
- Peters U, Jiao S, Schumacher FR, et al.: Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 144 (4): 799-807.e24, 2013. [PUBMED Abstract]
- Kastrinos F, Kupfer SS, Gupta S: Colorectal Cancer Risk Assessment and Precision Approaches to Screening: Brave New World or Worlds Apart? Gastroenterology 164 (5): 812-827, 2023. [PUBMED Abstract]
- Sassano M, Mariani M, Quaranta G, et al.: Polygenic risk prediction models for colorectal cancer: a systematic review. BMC Cancer 22 (1): 65, 2022. [PUBMED Abstract]
- Pearlman R, Frankel WL, Swanson B, et al.: Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol 3 (4): 464-471, 2017. [PUBMED Abstract]
- Boursi B, Sella T, Liberman E, et al.: The APC p.I1307K polymorphism is a significant risk factor for CRC in average risk Ashkenazi Jews. Eur J Cancer 49 (17): 3680-5, 2013. [PUBMED Abstract]
- Laken SJ, Petersen GM, Gruber SB, et al.: Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet 17 (1): 79-83, 1997. [PUBMED Abstract]
- Lothe RA, Hektoen M, Johnsen H, et al.: The APC gene I1307K variant is rare in Norwegian patients with familial and sporadic colorectal or breast cancer. Cancer Res 58 (14): 2923-4, 1998. [PUBMED Abstract]
Major Genetic Syndromes
Introduction
Originally described in the 1800s and 1900s by their clinical findings, the colon cancer susceptibility syndrome names often reflected the physician or patient and family associated with the syndrome (e.g., Gardner syndrome, Turcot syndrome, Muir-Torre syndrome, Lynch syndrome, Peutz-Jeghers syndrome [PJS], Bannayan-Riley-Ruvalcaba syndrome, and Cowden syndrome). These syndromes were associated with an increased lifetime risk of colorectal adenocarcinoma. They were mostly thought to have autosomal dominant inheritance patterns. Adenomatous colonic polyps were characteristic of the first four, while hamartomas were found to be characteristic in the last three.
With the development of the Human Genome Project and the identification in 1990 of the adenomatous polyposis coli (APC) gene on chromosome 5q, overlap and differences between these familial syndromes became apparent. Gardner syndrome and familial adenomatous polyposis (FAP) were shown to be synonymous, both caused by pathogenic variants in the APC gene. Attenuated FAP (AFAP) was recognized as a syndrome with less adenomas and extraintestinal manifestations due to an APC pathogenic variant at the 3’ or 5’ ends of the gene. MUTYH-associated polyposis (MAP) was recognized as a separate adenomatous polyp syndrome with autosomal recessive inheritance. Once the pathogenic variants were identified, the absolute risk of colorectal cancer (CRC) could be better assessed for carriers of pathogenic variants (refer to Table 3).
| Syndrome | Absolute Risk of CRC in Carriers of a Pathogenic Variant |
|---|---|
| FAP = familial adenomatous polyposis; JPS = juvenile polyposis syndrome; PJS = Peutz-Jeghers syndrome. | |
| aCancer risk estimates quoted here predate the widespread use of surveillance and prophylactic surgery. | |
| FAP a | 90% by age 45 y [1] |
| Attenuated FAP | 69% by age 80 y [2] |
| Lynch syndrome | 10% to 56% by age 75 y, depending on the gene involved [3-6] |
| MUTYH-associated polyposis | 35% to 53% [7] |
| PJS | 39% by age 70 y [8] |
| JPS | 17% to 68% by age 60 y [9,10] |
With these discoveries genetic testing and risk management became possible. Genetic testing refers to searching for variants in known cancer susceptibility genes using a variety of techniques. Comprehensive genetic testing includes sequencing the entire coding region of a gene, the intron-exon boundaries (splice sites), and assessment of rearrangements, deletions, or other changes in copy number (with techniques such as multiplex ligation-dependent probe amplification [MLPA] or Southern blot). Despite extensive accumulated experience that helps distinguish pathogenic variants from benign variants and polymorphisms, genetic testing sometimes identifies variants of uncertain significance (VUS) that cannot be used for predictive purposes.
Familial Adenomatous Polyposis (FAP)
Introduction
By 1900, several reports had demonstrated that patients with a large number of polyps (later subclassified as adenomas) were at very high risk of CRC and that the pattern of transmission in families was autosomal dominant. In the 20th century, the adenoma-to-carcinoma progression was confirmed, and FAP was recognized as the prototypical model for this progression.[11] Classic FAP is characterized by numerous (hundreds to thousands) adenomatous polyps in the colon and rectum developing after the first decade of life (refer to Figure 3).
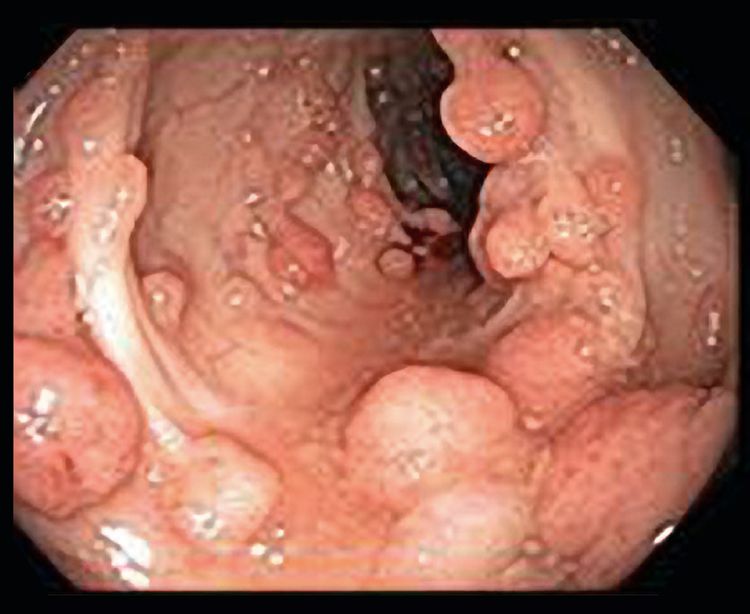
There is also a subset of classic FAP that has an attenuated phenotype. AFAP is a heterogeneous clinical entity characterized by fewer adenomatous polyps in the colon and rectum than in classic FAP. (Refer to the Attenuated Familial Adenomatous Polyposis [AFAP] section of this summary for more information.)
FAP is one of the most clearly defined and well understood of the inherited colon cancer syndromes.[1,12,13] It is an autosomal dominant condition, and the reported incidence varies from 1 in 7,000 to 1 in 22,000 live births.[14] The presence of ethnic differences in the prevalence of FAP has been suggested [14] but a large study did not find significant differences in ethnic variation in more than 6,169 individuals with a personal and/or family history of CRC and polyps who were referred for genetic testing at a large reference laboratory.[15] Most cases of FAP result from pathogenic variants in the APC gene on chromosome 5q21. (Refer to the Genetics of FAP section of this summary for more information about the APC gene and genetic testing.)
In addition to a high risk of colon adenomas in FAP patients, various extracolonic manifestations have also been described, including upper gastrointestinal (GI) tract adenomas and adenocarcinomas; fundic gland stomach polyps; nonepithelial benign tumors (osteomas, epidermal cysts, dental abnormalities); desmoid tumors; congenital hypertrophy of retinal pigment epithelium (CHRPE); and malignant tumors (thyroid and brain tumors, hepatoblastoma). Refer to Table 4 for the risks of these extracolonic manifestations in FAP.
| Malignancy | Relative Risk | Absolute Lifetime Risk (%) |
|---|---|---|
| Adapted from Giardiello et al.,[16] Jagelman et al.,[17] Sturt et al.,[18] Lynch et al.,[19] Bülow et al.,[20] Burt et al.,[21] and Galiatsatos et al.[22] | ||
| aThe Leeds Castle Polyposis Group. | ||
| Desmoid tumor | 852.0 | 15.0 |
| Duodenal tumors and cancer | 330.8 | 5.0–12.0 |
| Thyroid cancer | 7.6 | 2.0 |
| Brain cancer | 7.0 | 2.0 |
| Ampullary cancer | 123.7 | 1.7 |
| Pancreatic cancer | 4.5 | 1.7 |
| Hepatoblastoma | 847.0 | 1.6 |
| Gastric cancer | Not defined | 0.6a |
FAP has also been known as familial polyposis coli or adenomatous polyposis coli (APC). Gardner syndrome was previously the diagnosis for FAP patients who manifested with colorectal polyposis, osteomas, and soft tissue tumors. However, Gardner syndrome has been shown genetically to be a variant of FAP, and thus the term Gardner syndrome is essentially obsolete in clinical practice.[23]
Clinical phenotype
Colon adenomas and CRC
Individuals who inherit a pathogenic variant in the APC gene have a very high likelihood of developing colonic adenomas; the risk has been estimated to be more than 90%.[1,12,13] The age at onset of adenomas in the colon is variable, and the median age for the appearance of colorectal adenomas is 16 years.[24] By age 10 years, only 15% of carriers of the APC germline variant manifest adenomas; by age 20 years, the probability rises to 75%; and by age 30 years, 90% will have presented with FAP.[1,12,13,24,25] The exception is AFAP, in which affected individuals typically have fewer colon polyps, which are predominantly in the right colon, and later onset of CRC. (Refer to the Attenuated Familial Adenomatous Polyposis [AFAP] section of this summary for more information.) Without any intervention, most individuals with FAP will develop CRC by the fourth decade of life.[1,12,13] Thus, surveillance and intervention for carriers of an APC pathogenic variant and at-risk individuals have conventionally consisted of annual colonoscopy beginning around puberty for early detection of colonic polyps and to help plan when to perform colectomy.[26,27] (Refer to the Interventions for FAP section of this summary for more information.)
Extracolonic manifestations
Congenital hypertrophy of the retinal pigment epithelium (CHRPE)
CHRPE are flat, darkly pigmented lesions in the retina that are present in approximately 75% of patients with FAP [28,29] compared with a general population frequency of 1.2%.[30] The lesions are often present at birth or in early childhood and are frequently multiple or bilateral in FAP patients.[31] A study of 17 individuals diagnosed with FAP and 13 at-risk family members reported a sensitivity of the presence of a CHRPE lesion in association with colonic polyps in FAP of 76%, a specificity of 92%, a positive predictive value of 93%, and a negative predictive value of 75%; thus, screening at-risk individuals for CHRPE can be a reasonable method of detecting FAP.[28]
Desmoid tumors
Desmoid tumors are proliferative, locally invasive, nonmetastasizing, fibromatous tumors in a collagen matrix. Although they do not metastasize, they can grow very aggressively and be life threatening.[32] Desmoids may occur sporadically, as part of classical FAP, or in a hereditary manner without the colon findings of FAP.[19,33] Desmoids have been associated with hereditary APC pathogenic variants even when not associated with typical adenomatous polyposis of the colon.[33,34]
Most studies have found that 10% of FAP patients develop desmoids, with reported ranges of 8% to 38%. The incidence varies with the means of ascertainment and the location of the pathogenic variant in the APC gene.[33,35,36] APC pathogenic variants occurring between codons 1445 and 1578 have been associated with an increased incidence of desmoid tumors in FAP patients.[34,37-39] Desmoid tumors with a late onset and a milder intestinal polyposis phenotype (hereditary desmoid disease) have been described in patients with pathogenic variants at codon 1924.[33]
A desmoid risk factor scale has been described in an attempt to identify patients who are likely to develop desmoid tumors.[40] The desmoid risk factor scale was based on gender, presence or absence of extracolonic manifestations, family history of desmoids, and genotype, if available. By utilizing this scale, it was possible to stratify FAP patients into low-, medium-, and high-risk groups for developing desmoid tumors. The authors concluded that the desmoid risk factor scale could be used for surgical planning. Validation of the risk factors comprising this scale was supported by a large, multiregistry, retrospective study from Europe.[41]
The natural history of desmoids is variable. Some authors have proposed a model for desmoid tumor formation whereby abnormal fibroblast function leads to mesenteric, plaque-like desmoid precursor lesions, which in some cases occur before surgery and progress to mesenteric fibromatosis after surgical trauma, ultimately giving rise to desmoid tumors.[42] It is estimated that 10% of desmoids resolve, 50% remain stable for prolonged periods, 30% fluctuate, and 10% grow rapidly.[43] Desmoids often occur after surgical or physiological trauma, and both endocrine and genetic factors have been implicated. Approximately 80% of intra-abdominal desmoids in FAP occur after surgical trauma.[44,45]
The desmoids in FAP are often intra-abdominal, may present early, and can lead to intestinal obstruction or infarction and/or obstruction of the ureters.[36] In some series, desmoids are the second most common cause of death after CRC in FAP patients.[46,47] A staging system has been proposed to facilitate the stratification of intra-abdominal desmoids by disease severity.[48] The proposed staging system for intra-abdominal desmoids is as follows: stage I for asymptomatic nongrowing desmoids; stage II for symptomatic nongrowing desmoids of 10 cm or less in maximum diameter; stage III for symptomatic desmoids of 11 cm to 20 cm or for asymptomatic slow-growing desmoids; and stage IV for desmoids larger than 20 cm, or rapidly growing, or with life-threatening complications.[48]
These data suggest that genetic testing could be of value in the medical management of patients with FAP and/or multiple desmoid tumors. Those with APC genotypes predisposing to desmoid formation (e.g., at the 3’ end or codon 1445 of the APC gene) appear to be at high risk of developing desmoids after any surgery, including risk-reducing colectomy and surgical surveillance procedures such as laparoscopy.[35,43,49]
Stomach tumors
The most common FAP-related gastric polyps are fundic gland polyps (FGPs). FGPs are often diffuse and not amenable to endoscopic removal. The incidence of FGPs has been estimated to be as high as 60% in patients with FAP, compared with 0.8% to 1.9% in the general population.[20,22,50-54] These polyps consist of distorted fundic glands containing microcysts lined with fundic-type epithelial cells or foveolar mucous cells.[55,56]
The hyperplastic surface epithelium is, by definition, nonneoplastic. Accordingly, FGPs have not been considered precancerous. However, case reports of stomach cancer appearing to arise from FGPs have led to a reexamination of this issue.[22,57] In one FAP series, focal dysplasia was evident in the surface epithelium of FGPs in 25% of patients versus 1% of sporadic FGPs.[56] In a prospective study of patients with FAP undergoing surveillance with esophagogastroduodenoscopy, FGPs were detected in 88% of the patients. Low-grade dysplasia was detected in 38% of these patients, whereas high-grade dysplasia was detected in 3% of these patients. The study's authors recommended that, if a polyp with high-grade dysplasia is identified, polypectomy be considered with repeat endoscopic surveillance in 3 to 6 months.[58]
Complicating the issue of differential diagnosis, FGPs have been increasingly recognized in non-FAP patients consuming proton pump inhibitors (PPIs).[56,59] FGPs in this setting commonly show a PPI effect consisting of congestion of secretory granules in parietal cells, leading to irregular bulging of individual cells into the lumen of glands. To the trained eye, the presence of dysplasia and the concomitant absence of a characteristic PPI effect can be considered highly suggestive of the presence of underlying FAP. The number of FGPs tends to be greater in FAP than that seen in patients consuming PPIs, although there is some overlap.
Gastric adenomas also occur in patients with FAP. The incidence of gastric adenomas in Western patients is reported to be between 2% and 12%, whereas in Japan, incidence is reported to be between 39% and 50%.[60-63] These adenomas can progress to carcinoma. Patients with FAP in Korea and Japan have a threefold to fourfold increased risk of gastric cancer when compared with individuals in the general population in these countries. This finding was not observed in Western populations.[64-68] The increased prevalence of gastric adenomas in Asian patients with FAP may be due to the high prevalence of Helicobacter pylori infections in this population.[61]
More recently, a rise in incidence of gastric adenocarcinoma was observed in a Western FAP database.[69] Alterations in the promoter (1B) of APC were discovered in families with gastric adenocarcinoma and proximal polyposis of the stomach (GAPPS), who express numerous, predominantly fundic gland, gastric polyps restricted to the body and fundus with regions of dysplasia or gastric adenocarcinoma, and no evidence of colorectal or duodenal polyposis. These variants segregated with the gastric phenotype in multiple GAPPS families. Although penetrance of the gastric polyposis phenotype is high, the phenotype can vary ranging from asymptomatic adults to teenagers presenting with massive symptomatic gastric polyposis, as well as unaffected carriers who had clean endoscopies at ages ranging from 42 to 77 years. However, the penetrance for gastric cancer is less clear. Promoter 1B APC alterations rarely occur in FAP families with gastric fundic gland polyps and colonic polyposis.[70]
Duodenum/small bowel tumors
Whereas the incidence of duodenal adenomas is only 0.4% in unselected patients undergoing upper GI endoscopy,[71] duodenal adenomas are found in 80% to 100% of FAP patients. Most are located in the first and second portions of the duodenum, especially in the periampullary region.[50,51,72] In a prospective multicenter surveillance study of duodenal adenomas in 368 participants from northern Europe with FAP, 65% had adenomas at baseline evaluation (mean age, 38 y), with cumulative prevalence reaching 90% by age 70 years. In contrast to earlier beliefs regarding an indolent clinical course, the adenomas increased in size and degree of dysplasia during the 8 years of average surveillance, although only 4.5% developed cancer while under prospective surveillance.[20] This is a large study; however, it is limited by the use of forward-viewing rather than side-viewing endoscopy and the large number of investigators involved in the study. Intestinal polyps can also be assessed in FAP patients using capsule endoscopy.[73-75] One study of computed tomography (CT) duodenography found that larger adenoma size could be accurately measured but smaller, flatter adenomas could not be accurately counted.[76]
Approximately 1.7% of all duodenal cancer cases are attributed to FAP.[17,66,77-79] The incidence of duodenal adenocarcinoma in FAP is 4% to 12% lifetime. A retrospective review of FAP patients suggested that the adenoma-carcinoma sequence occurred in a temporal fashion for periampullary adenocarcinomas with a diagnosis of adenoma at a mean age of 39 years, high-grade dysplasia at a mean age of 47 years, and adenocarcinoma at a mean age of 54 years.[80] A decision analysis of 601 FAP patients suggested that the benefit of periodic surveillance starting at age 30 years led to an increased life expectancy of 7 months.[77] Although polyps in the duodenum can be difficult to treat, small series suggest that they can be managed successfully with endoscopy but with potential morbidity—primarily from pancreatitis, bleeding, and duodenal perforation.[81,82]
FAP patients with particularly severe duodenal polyposis, sometimes called dense polyposis, or with histologically advanced duodenal adenomas appear to be at the highest risk of developing duodenal adenocarcinoma.[20,78,83,84] Because the risk of duodenal adenocarcinoma is correlated with the number and size of polyps and the severity of dysplasia of the polyps, a stratification system that incorporates these features was developed to attempt to identify those individuals with FAP at the highest risk of developing duodenal adenocarcinoma.[84] According to this system, known as the Spigelman classification (refer to Table 5), 36% of patients with the most advanced stage will develop carcinoma.[78]
The predictive utility of the Spigelman classification has been called into question. The point system for dysplasia classifies dysplasia as mild, moderate, or severe, yet pathologists do not customarily attempt to distinguish moderate dysplasia from low-grade. There are no studies validating interobserver concordance in classifying a villous component or interpretation of the degree of dysplasia. A study from the Cleveland Clinic comparing Spigelman classification and its components in patients with FAP with and without cancer found neither adenoma count nor villous component to be predictive of cancer risk.[85] While patients with advanced (Spigelman IV) classification were more likely to develop cancer, nearly half of those with cancer did not meet Spigelman IV criteria.
| Points | Polyp Number | Polyp Size (mm) | Histology | Dysplasia |
|---|---|---|---|---|
| Stage I, 1–4 points; Stage II, 5–6 points; Stage III, 7–8 points; Stage IV, 9–12 points.[84] | ||||
| 1 | 1–4 | 1–4 | Tubular | Mild |
| 2 | 5–20 | 5–10 | Tubulovillous | Moderate |
| 3 | >20 | >10 | Villous | Severe |
Other tumors
Other extracolonic tumors arising in FAP patients include papillary thyroid cancer, adrenal tumors, hepatoblastoma, and brain tumors.
Papillary thyroid cancer (cribriform morular type) has been reported to affect 1% to 2% of patients with FAP.[86] However, a study [87] of papillary thyroid cancers in six women with FAP failed to demonstrate loss of heterozygosity (LOH) or pathogenic variants of the wild-type allele in codons 545 and 1061 to 1678 of the six tumors. In addition, four of five of these patients had detectable somatic RET/PTC chimeric genes. This pathogenic variant is generally restricted to sporadic papillary thyroid carcinomas, suggesting the involvement of genetic factors other than APC pathogenic variants. Further studies are needed to show whether other genetic factors such as the RET/PTC chimeric gene are independently responsible for or cooperative with APC variants in causing papillary thyroid cancers in FAP patients.
Adrenal tumors have been reported in FAP patients, and one study demonstrated LOH at the APC locus in an adrenocortical carcinoma (ACC) in an FAP patient.[88] In a study of 162 FAP patients who underwent abdominal CT for evaluation of intra-abdominal desmoid tumors, 15 patients (11 women) were found to have adrenal tumors.[89] Of these, two had symptoms attributable to cortisol hypersecretion. Three of these patients underwent subsequent surgery and were found to have ACC, bilateral nodular hyperplasia, or adrenocortical adenoma. The prevalence of an unexpected adrenal neoplasia in this cohort was 7.4%, which compares with a prevalence of 0.6% to 3.4% (P < .001) in non-FAP patients.[89] No molecular genetic analyses were provided for the tumors resected in this series. A subsequent study identified adrenal lesions in 26% (23 of 90) of patients with FAP, 18% (2 of 11) of patients with AFAP, and 24% (5 of 21) of patients with MAP. Most lesions in this series followed a benign and slowly progressive course; no cases of ACC were reported.[90]
Hepatoblastoma is a rare, rapidly progressive, and usually fatal childhood malignancy that, if confined to the liver, can be cured by radical surgical resection. Multiple cases of hepatoblastoma have been described in children with an APC pathogenic variant.[91-100] Some series have also demonstrated LOH of APC in these tumors.[92,94,101] No specific genotype-phenotype correlations have been identified in FAP patients with hepatoblastoma.[102] (Refer to the Hepatoblastoma section in the PDQ summary on Childhood Liver Cancer Treatment for more information.)
The constellation of CRC and brain tumors has been referred to as Turcot syndrome; however, Turcot syndrome is molecularly heterogeneous. Molecular studies have demonstrated that colon polyposis and medulloblastoma are associated with pathogenic variants in APC (thus representing FAP), while colon cancer and glioblastoma are associated with pathogenic variants in mismatch repair (MMR) genes (thus representing Lynch syndrome).[103]
Medulloblastoma, a highly malignant embryonal central nervous system tumor, accounts for approximately 80% of the brain tumors found in FAP and primarily occurs in children with 70% diagnosed before age 16 years. High-grade astrocytomas and ependymomas have also been described in FAP patients. Although the relative lifetime risk of any brain tumor among members of an FAP family is increased 7-fold and that of medulloblastoma 90-fold, the absolute lifetime risk of any brain tumor is approximately 1% to 2%.[103]
Genetics of FAP
APC gene
The APC gene on chromosome 5q21 encodes a 2,843-amino acid protein that is important in cell adhesion and signal transduction; the main function of the APC protein is to regulate intracellular concentrations of beta-catenin, a major mediator of the Wnt signal transduction pathway. APC is a tumor suppressor gene, and the loss of APC is among the earliest events in the chromosomal instability colorectal tumor pathway. FAP and AFAP can be diagnosed genetically by testing for germline pathogenic variants in the APC gene in DNA from peripheral blood leukocytes. More than 300 different disease-associated pathogenic variants of the APC gene have been reported.[104] Most of these changes are insertions, deletions, and nonsense variants that lead to frameshifts and/or premature stop codons in the resulting transcript of the gene. The most common APC pathogenic variant (10% of FAP patients) is a deletion of AAAAG in codon 1309; no other pathogenic variants appear to predominate. Variants that reduce rather than eliminate production of the APC protein may also lead to FAP.[105]
Genotype-phenotype correlations
Most APC pathogenic variants that occur between codon 169 and codon 1249 result in the classic FAP phenotype.[106-108] There has been much interest in correlating the location of the pathogenic variant within the gene with the clinical phenotype:
- Researchers have found that dense carpeting of colonic polyps, a feature of classic FAP, is seen in most patients with APC pathogenic variants, particularly those variants that occur between codons 1250 and 1464. AFAP is associated with pathogenic variants that occur in or upstream of exon 4 and in the latter two-thirds of exon 15.[106-109] (Refer to the Attenuated Familial Adenomatous Polyposis [AFAP] section of this summary for more information.)
- CHRPE are rarely associated with pathogenic variants that occur before exon 9.[39,108] Individuals with exon 9 variants tend not to have duodenal adenomas.[70,110]
- Families with GAPPS, who express numerous, predominantly fundic gland gastric polyps restricted to the body and fundus with regions of dysplasia or gastric adenocarcinoma, and no evidence of colorectal or duodenal polyposis, were found to possess variants in the promoter (1B) of APC.[70]
- APC pathogenic variants occurring between codons 1445 and 1578 have been associated with an increased incidence of desmoid tumors in FAP patients.[34,37-39]
A low-penetrance APC variant, I1307K, has been studied for its association with CRC. (Refer to the APC I1307K section in the Colorectal Cancer Susceptibility Genes section of this summary for more information.)
Genetic testing for FAP
Probands
Individuals who present with a classic FAP phenotype are candidates for APC testing. However, in many probands with a personal or family history of polyposis, multigene panel testing is an appropriate option to consider given the genetic heterogeneity of polyposis conditions and the phenotypic overlap among associated syndromes.
In particular, patients who develop fewer than 100 colorectal adenomatous polyps may pose a diagnostic challenge. The differential diagnosis includes AFAP, MAP, polymerase proofreading–associated polyposis (PPAP), and constitutional mismatch repair deficiency (CMMRD).[111] AFAP can be diagnosed by testing for germline APC pathogenic variants. (Refer to the Attenuated Familial Adenomatous Polyposis [AFAP] section of this summary for more information.) MAP is caused by biallelic germline pathogenic variants in the MUTYH gene, inherited in an autosomal recessive manner.[112] PPAP is caused by heterozygous pathogenic variants in POLE and POLD1.[113,114] CMMRD is a condition in which individuals inherit pathogenic variants in both alleles of one MMR gene (MLH1, MSH2, MSH6, PMS2, or EPCAM).[115] For more information, see the MUTYH-Associated Polyposis (MAP), Oligopolyposis, and IHC in constitutional mismatch repair deficiency (CMMRD) syndrome sections.
For example, in a large cross-sectional study, pathogenic variants in APC were found in 80% (95% confidence interval [CI], 71%–87%) of individuals with more than 1,000 adenomas, 56% (95% CI, 54%–59%) in those with 100 to 999 adenomas, 10% (95% CI, 9%–11%) in those with 20 to 99 adenomas, and 5% (95% CI, 4%–7%) in those with 10 to 19 adenomas.[116] In this same study, the prevalence of biallelic MUTYH pathogenic variants was similar to APC for those with the attenuated phenotype (20–99 adenomas), but MUTYH pathogenic variants were also observed in a small minority (2%) of those with classic polyposis.[116]
Most commercial laboratories perform not only full gene sequencing but also deletion/duplication analysis of the APC and other genes. However, it is important to verify the testing methodology with each laboratory. Deletion analysis is especially important for individuals with FAP because 8% to 12% of affected individuals have a whole exon deletion or promoter 1B deletion in the APC gene, which would not be detected with sequencing.[117-120] As mentioned, for patients who present with polyposis, multigene panels that include multiple polyposis genes are often ordered, which simplifies and lowers the cost of testing by assessing all genes at the same time. (Refer to the Multigene [panel] testing section in the PDQ summary on Cancer Genetics Risk Assessment and Counseling for more information.)
Cascade testing
In families in which a pathogenic variant in the APC gene is identified, predictive testing for at-risk relatives can definitively identify or rule out the variant. Such testing is important to determine whether at-risk relatives need to undergo aggressive screening or whether such procedures are not necessary or can be discontinued (i.e., in relatives who test negative for the familial pathogenic variant).
Most patients with FAP have an affected parent, and a pattern of autosomal dominant inheritance may be observed in the family. Accordingly, cascade genetic counseling and testing may then be extended to at-risk family members. However, it is estimated that 25% of patients with FAP have a de novo pathogenic variant in APC, meaning that the variant does not appear to be inherited from either parent.[121] In cases where the variant cannot be identified in leukocyte DNA of either parent, it is possible that germline mosaicism may explain the finding. Thus, siblings of an individual should always be offered APC testing, but testing aunts, uncles, and cousins of the proband would not be indicated.
The early appearance of FAP clinical features and the subsequent recommendations for surveillance beginning at puberty raise special considerations relating to the genetic testing of minors.[122] Hereditary cancer genetic testing is not recommended for minors unless results will affect medical management in childhood. Thus, FAP presents an example in which possible medical benefit justifies genetic testing of minors in families with a known pathogenic variant, especially for the anticipated 50% of at-risk children who will be found not to be carriers of pathogenic variants and who can thus be spared surveillance. In addition, testing infants for FAP can allow for hepatoblastoma surveillance until age 5 years. Otherwise, if at-risk minors are not tested, high-quality colonoscopy is initiated between ages 10 and 15 years.[123] The psychological impact of such testing is addressed in the Psychosocial Issues in Hereditary Colon Cancer Syndromes section of this summary.
Interventions for FAP
Colon surveillance
Individuals at risk of FAP, because of a known APC pathogenic variant in either the family or themselves, are evaluated for onset of polyposis by flexible sigmoidoscopy or colonoscopy. Once an FAP family member is found to manifest polyps, the only effective management to prevent CRC is colectomy. Prophylactic surgery has been shown to improve survival in patients with FAP.[124] If feasible, the patient and his/her family members should be included in a registry because it has been shown retrospectively that registration and surveillance reduce CRC incidence and mortality.[125] In patients with classic FAP identified very early in their course, the surgeon, endoscopist, and family may choose to delay surgery for several years in the interest of achieving social milestones. In addition, in carefully selected patients with AFAP (those with minimal polyp burden and advanced age), deferring a decision about colectomy may be reasonable with surgery performed only in the face of advancing polyp burden or dysplasia.
A Finnish nationwide, population-based, retrospective study evaluating whether surveillance of family members with FAP reduced overall mortality and improved survival demonstrated that family members of probands who were recruited to the screening program had equivalent survival to the general population up to 20 years after diagnosis of FAP.[126] The study included 154 families with at least one family member clinically diagnosed with FAP from 1963 to 2015. There were 194 probands and 225 family members (83 diagnosed by genetic testing and 142 by endoscopy) with a median time of follow-up of 11.8 years. In this study, the survival analysis of members of FAP families was calculated using the relative survival estimate.[127] This estimate compares survival among FAP probands and family members with the survival expected in the absence of FAP among individuals of the same gender and age in each calendar year. The relative survival rate was 67% for probands (95% CI, 60%–75%) at 10 years of follow-up and 66% (95% CI, 58%–76%) at 20 years of follow-up. For family members, the relative survival rate was 98% (95% CI, 95%–101%) at 10 years of follow-up and 94% (95% CI, 88%–100%) at 20 years of follow-up. The relative survival rate was 87% (95% CI, 79%–96%) for family members at 25 years of follow-up. The relative survival rate for probands was significantly lower than the relative survival rate for family members (P < .001). The standardized mortality ratio was elevated in probands in both the 0- to 5-year and 5- to 10-year periods of follow-up whereas it remained stable for family members until 20 years of follow-up. This difference was more marked in the beginning of follow-up for probands, considering that most of them were probably symptomatic and most likely had CRC when they were diagnosed with FAP. The authors mentioned that if the CRC was treated successfully without recurrence, the survival of the probands approached that of the family members.
Colonoscopic surveillance usually begins at an early age (10–15 y) in individuals with FAP.[123] (Refer to the Psychosocial Issues in Hereditary Colon Cancer Syndromes section of this summary for more information on the social and emotional implications of early surveillance.) Colonoscopy is the screening tool of choice for individuals with FAP for the following reasons: (a) improved instrumentation for full colonoscopy, (b) sedation, (c) recognition of AFAP, in which the disease typically manifests in the right colon, and (d) the growing tendency to defer surgery for a number of years.[123] The National Comprehensive Cancer Network (NCCN) recommends that individuals who test negative for a known familial APC pathogenic variant undergo average-risk population screening. However, clinical surveillance is warranted in families in which an APC pathogenic variant has not been identified in an affected individual. NCCN also recommends that individuals with an APC pathogenic variant continue colon cancer screening, even if they have not developed colon polyps, since adenomas may not manifest until later in life.[123] (Refer to the PDQ summary on Colorectal Cancer Screening for more information on these methods.)
Colorectal surgery
Colon adenomas will develop in nearly 100% of individuals who are APC pathogenic variant positive; risk-reducing surgery comprises the standard of care to prevent CRC after polyps have appeared and are too numerous or histologically advanced to monitor safely using endoscopic resection.
FAP patients and their doctors should have an individualized discussion to decide when surgery will be performed. It is useful to incorporate into the discussion the risk of developing desmoid tumors after surgery, as well as fecundity for women. Timing of risk-reducing surgery usually depends on the number of polyps, their size, histology, and symptomatology.[128] Once numerous polyps have developed, surveillance colonoscopy is no longer useful in timing the colectomy because polyps are so numerous that it is not possible to biopsy or remove all of them. At this time, it is appropriate for patients to consult with a surgeon who is experienced with available options, including total colectomy and restorative proctocolectomy.[129] Rectum-sparing surgery, with sigmoidoscopic surveillance of the remaining rectum, is a reasonable alternative to total colectomy in those compliant individuals with relative rectal sparing of polyps who understand the consequences and make an informed decision to accept the residual risk of rectal cancer occurring despite periodic surveillance.[130]
Surgical options include restorative proctocolectomy with ileal pouch–anal anastomosis (IPAA), total colectomy with ileorectal anastomosis (IRA), or total proctocolectomy with ileostomy (TPC). TPC is reserved for patients with low rectal cancer in which the sphincter cannot be spared or for patients on whom an IPAA cannot be performed because of technical problems. There is no risk of developing rectal cancer after TPC because the whole mucosa at risk is removed. These procedures can be performed using minimally invasive techniques.
Irrespective of whether a colectomy and an IRA or a restorative proctocolectomy is performed, most experts suggest that periodic and lifelong surveillance of the rectum or the ileal pouch be performed to remove or ablate any polyps. In earlier unselected studies, the risk of rectal cancer after total colectomy 20 years after IRA was reported to be as high as 25%.[131,132] This risk has been reported to be much lower with better selection of patients for IRA.[129,133] Factors that have been reported to increase rectal cancer risk after IRA include the number of polyps throughout the colon, the number of polyps in the rectum, the presence of colon cancer at the time of IRA, the length of the rectal stump, the duration of follow-up after IRA, and the genotype.[39,134-136] An abdominal colectomy with IRA as the primary surgery for FAP does not preclude later conversion to an IPAA for uncontrolled rectal polyps and/or rectal cancer. In the Danish Polyposis Registry, the morbidity and functional results of a secondary IPAA (after a previous IRA) in 24 patients were reported to be similar to those of 59 patients who underwent primary IPAA.[137]
In most cases, the clinical polyp burden in the rectum at the time of surgery dictates the type of surgical intervention, namely, restorative proctocolectomy with IPAA versus IRA. Patients with a mild phenotype (<1,000 colonic adenomas) and fewer than 20 rectal polyps may be candidates for IRA at the time of prophylactic surgery.[138] In some cases, however, the polyp burden is equivocal, and in such cases, investigators have considered the role of genotype in predicting subsequent outcomes with respect to the rectum.[139] Several APC pathogenic variants can increase an individual's rectal cancer risk and can therefore increase an individual's risk for eventual completion proctectomy after IRA. These APC pathogenic variants are located in exon 15 in codon 1250, codon 1309, codon 1328, and between codons 1250 and 1464.[140,131,141,142] A meta-analysis examined quality of life after restorative proctocolectomy and IPAA in patients with ulcerative colitis and FAP. The results suggested that patients with FAP do marginally better than patients with ulcerative colitis regarding fistula formation, pouchitis, stool frequency, and seepage.[143]
It is important to continue annual surveillance of the ileal pouch in patients who have undergone IPAA because they are at risk of developing neoplasia in the anal transitional zone/residual rectal mucosa and in the ileal pouch. The cumulative risk of developing adenomas in the ileal pouch can be up to 75% for 15 years after surgery has been completed.[144,145] A retrospective study from the Cleveland Clinic Registry followed patients with FAP who underwent IPAA. After a median follow-up period of 10.1 years and a median of four pouchoscopies, 47% of patients (78 of 165) developed rectal mucosa/transitional zone adenomas. Adenomas were more frequently diagnosed in patients who had undergone stapled IPAA (52.3% [72 of 137]) when compared with those who had undergone mucosectomy and handsewn IPAA (21.4% [6 of 28]; P < .005).[146] Although they are rare, carcinomas have been reported in the ileal pouch and anal transition zone after restorative proctocolectomy in patients with FAP.[147] In the Cleveland Clinic study, six patients developed cancer after IPAA (three patients developed cancer while under surveillance, and three patients developed cancer after missing surveillance pouchoscopies).[146]
Chemoprevention
Celecoxib, a specific cyclooxygenase 2 (COX-2) inhibitor, and nonspecific COX-2 inhibitors, such as sulindac (a nonsteroidal anti-inflammatory drug [NSAID]), have been associated with a decrease in polyp size and number in FAP patients, suggesting a role for chemopreventive agents in the treatment of this disorder.[148,149] Although celecoxib had been approved by the U.S. Food and Drug Administration (FDA), its license was voluntarily withdrawn by the manufacturer. Currently, there are no FDA-approved drugs for chemoprevention in FAP. Nevertheless, agents such as celecoxib and sulindac are used so commonly that chemopreventive clinical trials typically use one of these agents as a control. A randomized trial showed possible marginal improvement in polyp burden with the combination of celecoxib and eflornithine (i.e., difluoromethylornithine [DFMO]) when compared with celecoxib alone.[150] An international randomized trial analyzed the use of daily sulindac, eflornithine, or sulindac plus eflornithine in 171 individuals with FAP. In the three study arms, there were no significant differences between the frequencies of FAP-related events (i.e., primary end points), which were composites of major surgeries (colectomies/proctocolectomies/pouchectomies/duodenal resections), excisions of advanced adenomas, diagnoses of cancer/high-grade dysplasia, and/or progressions of duodenal polyposis by more than one Spigelman stage.[151] A post-hoc analysis was performed on 158 participants with FAP who were at risk for lower GI neoplasia. Results demonstrated a significant, marked reduction in the likelihood of lower GI tract polyposis progression (hazard ratio [HR], <0.2) in the sulindac plus eflornithine arm versus sulindac alone (HR, 0.195; 95% CI, 0.048–0.803) and versus eflornithine alone (HR, 0.171; 95% CI, 0.042–0.698). Lower GI tract polyposis progression was defined as the need for colectomy, proctocolectomy, proctectomy, pouchectomy, the excision of any polyp that was more than 10 mm, and/or the diagnosis of cancer/high-grade dysplasia. Moreover, 0 of 54 participants randomly assigned to the sulindac plus eflornithine arm required major surgery, versus 7 of 53 (13.2%) in the sulindac-only arm, and 8 of 51 (15.7%) in the eflornithine-only arm (P ≤ .005).[152]
A small, randomized, placebo-controlled, dose-escalation trial of celecoxib in a pediatric population (aged 10–14 y) demonstrated the safety of celecoxib at all dosing levels when administered over a 3-month period.[153] This study found a dose-dependent reduction in adenomatous polyp burden. At a dose of 16 mg/kg/day, which approximates the approved dose of 400 mg twice daily in adults, the reduction in polyp burden paralleled that demonstrated with celecoxib in adults.
Omega-3-polyunsaturated fatty acid eicosapentaenoic acid in the free fatty acid form has been shown to reduce rectal polyp number and size in a small study of patients with FAP after subtotal colectomy.[154] Although not directly compared in a randomized trial, the effect appeared to be similar in magnitude to that previously observed with celecoxib.
It is unclear at present how to incorporate COX-2 inhibitors into the management of FAP patients who have not yet undergone risk-reducing surgery. A double-blind placebo-controlled trial of 41 child and young adult carriers of APC pathogenic variants who had not yet manifested polyposis demonstrated that sulindac may not be effective as a primary treatment in FAP. There were no statistically significant differences between the sulindac and placebo groups over 4 years of treatment in incidence, number, or size of polyps.[149]
Consistent with the effects of COX-2 inhibitors on colonic polyps, in a randomized, prospective, double-blind, placebo-controlled trial, celecoxib reduced, but did not eliminate, the number of duodenal polyps in 32 patients with FAP after a 6-month course of treatment. Of importance, a statistically significant effect was seen only in individuals who had more than 5% of the duodenum involved with polyps at baseline and with an oral dose of 400 mg, given twice daily.[155] A previous randomized study of 24 FAP patients treated with sulindac for 6 months showed a nonsignificant trend in the reduction of duodenal polyps.[156] The same issues surrounding the use of COX-2 inhibitors for the treatment of colonic polyps apply to their use for the treatment of duodenal polyps (e.g., only partial elimination of the polyps, complications secondary to the COX-2 inhibitors, and loss of effect after the medication is discontinued).[155]
Because of the common clustering of adenomatous polyps around the duodenal papilla (where bile enters the intestine) and preclinical data suggesting that ursodeoxycholate inhibits intestinal adenomas in mice that harbor an Apc germline variant,[157] two trials that employ ursodeoxycholate have been performed.[158,159] In both studies, ursodeoxycholate did not have a significant chemopreventive effect on duodenal polyps; paradoxically, in one study, ursodeoxycholate in combination with celecoxib appeared to promote polyp density in patients with FAP.
Because of reports demonstrating an increase in cardiac-related events in patients taking rofecoxib and celecoxib,[160-162] it is unclear whether this class of agents will be safe for long-term use for patients with FAP and in the general population. Also, because of the short-term (6 months) nature of these trials, there is currently no clinical information about cardiac events in FAP patients taking COX-2 inhibitors on a long-term basis.
Level of evidence (celecoxib): 1b
One cohort study has demonstrated regression of colonic and rectal adenomas with sulindac treatment in FAP. The reported outcome of this trial was the number and size of polyps, a surrogate for the clinical outcome of main interest, CRC incidence.[163]
Level of evidence (sulindac): 1b
Preclinical studies of a small-molecule epidermal growth factor receptor (EGFR) inhibitor and low-dose sulindac in the Apcmin/+ mouse diminished intestinal adenoma development by 87% [164] suggesting that EGFR inhibitors had the potential to inhibit duodenal polyps in FAP patients. A 6-month double-blind, randomized, placebo-controlled trial tested the efficacy of sulindac, 150 mg twice daily, and erlotinib, 75 mg daily, versus placebo in FAP or AFAP patients with duodenal polyps.[165] Ninety-two patients with FAP or AFAP were randomly assigned to receive study drugs or placebo and underwent pretreatment and posttreatment upper endoscopies to determine the changes in the sum diameter of the polyps and number of polyps in a 10 cm segment of proximal duodenum. The trial was terminated prematurely because the primary endpoint was met. The intent-to-treat analysis demonstrated a median decrease in duodenal polyp burden (sum of diameters) of 8.5 mm in the sulindac/erlotinib arm while there was an 8 mm increase in the placebo arm (P < .001). Significantly higher rates of grade 1 and grade 2 adverse events occurred in the treatment arm than in the placebo arm: in the treatment arm, 60.9% developed an acneiform rash and 32.6% developed oral mucositis; in the placebo arm, 19.6% developed an acneiform rash and 10.9% developed oral mucositis. A 2018 trial in 82 individuals with FAP found that combination treatment with sulindac (150 mg twice daily) and erlotinib (75 mg daily) compared with placebo resulted in a significantly lower (69.4%) colorectal polyp burden after 6 months of treatment (95% CI, 28.8%–109.2%; P = .009).[166] There was a reduction in polyp burden in both those with an entire colorectum and those with only a rectal pouch or rectum. However, it is unclear to what extent erlotinib contributed to this reduction given that sulindac has been proven effective in the lower GI tract.
On the basis of the previously modest effects of sulindac and celecoxib on duodenal polyps in patients with FAP [149,163] and the dramatic effect of genetic EGFR inhibition on intestinal adenoma development in the Apcmin/+ mouse,[167] it is likely that erlotinib was responsible for the success of these trials. An ongoing clinical trial (NCT02961374) is determining whether lower doses of erlotinib alone are sufficient for significantly reducing duodenal polyp burden in patients with FAP and AFAP.
Management of extracolonic tumors
Patients who carry APC germline pathogenic variants are at increased risk of other types of malignancies, including desmoid tumors, gastric tumors, duodenal cancer, small bowel cancer, hepatoblastoma, thyroid cancer, and brain tumors. The management of these extracolonic tumors is described below.
Desmoid tumors
The management of desmoids in FAP can be challenging and can complicate prevention efforts. There is no accepted standard treatment for desmoid tumors. Multiple medical treatments have generally been unsuccessful in the management of desmoids. Treatments have included antiestrogens, NSAIDs, chemotherapy, and radiation therapy, among others. Studies have evaluated the use of raloxifene alone, tamoxifen or raloxifene combined with sulindac, and pirfenidone alone.[168-170]
Thirteen patients with intra-abdominal desmoids and/or unfavorable response to other medical treatments who had expression of estrogen-alpha receptors in their desmoid tissues were included in a prospective study of raloxifene, given in doses of 120 mg daily.[168] Six patients had been on tamoxifen or sulindac before treatment with raloxifene, and seven patients were previously untreated. All 13 patients with intra-abdominal desmoid disease had either a partial or a complete response 7 months to 35 months after starting treatment, and most desmoids decreased in size at 4.7 months (± 1.8 mo) after treatment. Response occurred in patients with desmoid plaques and with distinct lesions. Study limitations include small sample size and the clinical evaluation of response, which was not consistent in all patients. Several questions remain concerning the outcomes of patients with desmoid tumors not expressing estrogen-alpha receptors who have received raloxifene, as well as which patients may benefit from this potential treatment.
A second study of 13 patients with FAP-associated desmoid tumors, who were treated with tamoxifen 120 mg/day or raloxifene 120 mg/day in combination with sulindac 300 mg/day, reported that ten patients had either stable disease (n = 6) or a partial or complete response (n = 4) for more than 6 months and that three patients had stable disease for more than 30 months.[169] These results suggest that the combination of these agents may be effective in slowing the growth of desmoid tumors. However, the natural history of desmoids is variable, with both spontaneous regression and variable growth rates.
A third study reported mixed results in 14 patients with FAP-associated desmoid tumors treated with pirfenidone for 2 years.[170] In this study, some patients had disease regression, some patients had disease progression, and some patients had stable disease.
There are reports of using imatinib mesylate to treat desmoid tumors in FAP patients with some success.[171,172] Nilotinib demonstrated potential to stabilize desmoid tumor growth after treatment failure with imatinib in patients with desmoid tumors.[173]
The benefit of the tyrosine kinase inhibitor sorafenib in the treatment of desmoid tumors was demonstrated in a phase III randomized trial comparing sorafenib (400 mg daily) with placebo in 87 patients with unresectable progressive or symptomatic desmoid tumors.[174] Crossover to the sorafenib group was permitted for patients in the placebo group who had disease progression on the placebo arm of the study. Objective responses were demonstrated in 16 of 49 patients treated with sorafenib (33%) compared with 7 of 35 placebo-treated patients (20%). Additionally, the 2-year progression-free survival (PFS) rate was significantly higher for sorafenib (81%) than placebo (36%); the HR for progression or death was 0.13 (95% CI, 0.05–0.31; P < .001). The most frequently reported adverse events were grade 1 or grade 2 rash (73%), fatigue (67%), hypertension (55%), and diarrhea (51%). Despite a relatively favorable toxicity profile, approximately 20% of patients discontinued sorafenib due to toxicity, emphasizing the importance of appropriate dose delays and interruptions for the treatment of adverse events.
Because of the high rates of morbidity and recurrence, in general, surgical resection is not recommended in the treatment of intra-abdominal desmoid tumors. A review of experiences at one hospital suggested that surgical outcomes with intra-abdominal desmoid tumors may be better than previously believed.[175,176] Issues of subject selection are critical in evaluating surgical outcome data.[175] Abdominal wall desmoid tumors can be treated with surgical resection, but the recurrence rate is high.
Stomach tumors
It is not clear how to manage gastric adenomas. Only retrospective case series are available and point to a relatively low prevalence of gastric adenocarcinoma development in FAP patients.[177,178] More recently, a rise in incidence of gastric adenocarcinoma was observed in a Western FAP database [69] suggesting that a possible change in the management of gastric tumorigenesis in FAP may be in order. One group recommends endoscopic polypectomy for the management of gastric adenomas.[69] The management of adenomas in the stomach is usually individualized based on the size of the adenoma and the degree of dysplasia.
Duodenum/small bowel tumors
Endoscopic surveillance usually begins between ages 20 to 25 years in patients with FAP. Baseline upper endoscopy may be performed at an earlier age if the patient has a family history of large duodenal adenoma burden or duodenal/ampullary cancer.[67] The subsequent intervals between endoscopy vary according to the findings of the previous endoscopy, often based on Spigelman stage. Recommended intervals are based on expert opinion although the relatively liberal intervals for stage 0 to stage II disease are based in part on the natural history data generated by the Dutch/Scandinavian duodenal surveillance trial (refer to Table 6 for available recommendations regarding screening frequency by Spigelman stage).[20]
The main advantages of the Spigelman classification are its long-standing familiarity to and usage by those in the field, which allows reasonable standardization of outcome comparisons across studies.[63,179] However, the following are limitations of application of the Spigelman classification:
- Most pathologists do not employ the term moderate dysplasia, preferring a simpler low- versus high-grade dysplasia system.
- Because of the villous nature of normal duodenal epithelium, pathologists commonly disagree over the classification of tubular, tubulovillous, and villous.
- Spigelman staging requires biopsy, which is not always essential when only a few small plaques are present; conversely, for larger adenomas, sampling variation leads to understaging.[180,181]
| Spigelman Stage | NCCN (2024) [123] | ESMO (2013) [182] |
|---|---|---|
| ESMO = European Society of Medical Oncology; NCCN = National Comprehensive Cancer Network. | ||
| See below for additional information about the use of surgical resection in Spigelman stage IV disease. | ||
| 0 (no polyps) | Endoscopy every 3–5 y | Not specified |
| I | Endoscopy every 2–3 y | Endoscopy every 5 y |
| II | Endoscopy every 1–2 y | Endoscopy every 3 y |
| III | Endoscopy every 6–12 mo | Endoscopy every 1–2 y |
| IV | Expert endoscopic surveillance every 3–6 mo | Endoscopy every 6-12 mo |
| Excision/ablation of resectable large or villous adenomatous polyps and endoscopic ampullectomy are options that may help individuals avoid surgery | ||
| Surgical evaluation and counseling for individuals with high-grade dysplasia, invasive carcinoma, or dense polyposis that cannot be removed endoscopically | Surgical options include duodenotomy with polypectomy, pancreas-sparing duodenectomy and pancreaticoduodenectomy (Whipple procedure) | |
The results of long-term duodenal adenoma surveillance of FAP patients in Nordic countries and the Netherlands revealed significant duodenal cancer risk in FAP patients.[183] According to the protocol, biennial frontal-viewing endoscopy was performed from 1990 through 2000. Subsequently, patients were followed up with surveillance according to international guidelines. The study group comprised 261 of 304 patients (86%) who had more than one endoscopy. Median follow-up was 14 years (range, 9–17 y). The lifetime risk of duodenal adenomatosis was 88%. Forty-four percent of patients had worsening Spigelman stage over time, whereas 12% improved and 34% remained unchanged. Twenty patients (7%) developed duodenal cancer at a median age of 56 years (range, 44–82 y). The cumulative cancer incidence was 18% at age 75 years (95% CI, 8%–28%). Survival in patients with symptomatic cancers was worse than those diagnosed at surveillance endoscopy.
Level of evidence (screening for duodenum/small bowel tumors): 3
Many factors, including severity of polyposis, comorbidities, patient preferences, and availability of adequately trained physicians, determine whether surgical or endoscopic therapy is selected for polyp management. Endoscopic resection or ablation of large or histologically advanced adenomas appears to be safe and effective in reducing the short-term risk of developing duodenal adenocarcinoma;[81,82,184] however, patients managed with endoscopic resection of adenomas remain at substantial risk of developing recurrent adenomas in the duodenum.[180] The most definitive procedure for reducing the risk of adenocarcinoma is surgical resection of the ampulla and duodenum, although these procedures also have higher morbidity and mortality associated with them than do endoscopic treatments. Duodenotomy and local resection of duodenal polyps or mucosectomy have been reported, but invariably, the polyps recur after these procedures.[185] In a series of 47 patients with FAP and Spigelman stage III or stage IV disease who underwent definitive radical surgery, the local recurrence rate was reported to be 9% after a mean follow-up period of 44 months. This local recurrence rate was dramatically lower than any local endoscopic or surgical approach from the same study.[180] Pancreaticoduodenectomy and pancreas-sparing duodenectomy are appropriate surgical therapies that are believed to substantially reduce the risk of developing periampullary adenocarcinoma.[181,185-187] If such surgical options are considered, preservation of the pylorus will benefit this patient group because most will have undergone a subtotal colectomy with IRA or total colectomy with IPAA. A matched case-control study of 32 people with FAP (all patients had pancreaticoduodenectomy after IPAA) found that pancreaticoduodenectomy did not impact quality of life.[188] As noted in a Northern European study,[20] and others,[189,190] most patients with duodenal adenomas will not develop cancer and can be followed with endoscopy. However, individuals with advanced adenomas (Spigelman stage III or stage IV disease) generally require endoscopic or surgical treatment of the polyps. Chemoprevention studies for duodenal adenomas in FAP patients are under way and may offer an alternate strategy in the future. (Refer to the Chemoprevention section of this summary for more information.)
The endoscopic approach to larger and/or flatter adenomas of the duodenum depends on whether the ampulla is involved. Endoscopic mucosal resection (EMR) after submucosal injection of saline, with or without epinephrine and/or dye, such as indigo carmine, can be employed for nonampullary lesions. Ampullary lesions require even greater care including endoscopic ultrasound evaluation for evidence of bile or pancreatic duct involvement. Stenting of the pancreatic duct is commonly performed to prevent stricturing and pancreatitis. The stents require endoscopic removal at an interval of 1 to 4 weeks. Because the ampulla is tethered at the ductal orifices, it typically does not uniformly lift with injection, so injection is commonly not used. Any consideration of EMR or ampullectomy requires great experience and judgment, with careful consideration of the natural history of untreated lesions and an appreciation of the high rate of adenoma recurrence despite aggressive endoscopic intervention.[82,180,181,186,191-194] The literature uniformly supports duodenectomy for Spigelman stage IV disease. For Spigelman stage II and stage III disease, there is a role for endoscopic treatment invariably focusing on the one or two worst lesions that are present.
Reluctance to consider surgical resection is related to the short-term morbidity and mortality and the long-term complications related to surgery. Although these concerns are likely overstated,[180,181,187,191,195-201] fear of surgical intervention can lead to aggressive and somewhat ill-advised endoscopic interventions. In some circumstances, endoscopic resection of ampullary and/or other duodenal adenomas cannot be accomplished completely or safely by endoscopic means, and duodenectomy cannot be accomplished without risking a short-gut syndrome or cannot be done at all because of mesenteric fibrosis. In such cases, surgical transduodenal ampullectomy/polypectomy can be performed. However, this is associated with a high risk of local recurrence similar to that of endoscopic treatment.
Level of evidence (treatment of duodenum/small bowel tumors): 4
Other tumors
Although level 1 evidence is lacking for the following surveillance methods, they are based on expert opinion. NCCN recommends baseline thyroid ultrasound beginning in the late teenage years to screen for papillary thyroid cancer in patients with FAP, with a repeat ultrasound every 2 to 5 years if results are normal. When individuals have a family history of thyroid cancer, shorter screening intervals can be used.[123,202,203]
Level of evidence (thyroid cancer ultrasound screening): 4
Although level 1 evidence is lacking for the following surveillance methods, they are based on expert opinion. NCCN has suggested that the following be considered for children with a predisposition to FAP: liver palpation, abdominal ultrasound, and measurement of serum alpha-fetoprotein every 3 to 6 months for the first 5 years of life.[123,204] It is not necessary to continue screening after age 5 years.
Level of evidence (hepatoblastoma or adrenal cancer screening): 5
Although level 1 evidence is lacking for the following surveillance methods, they are based on expert opinion. Medulloblastoma is a highly malignant tumor that is usually only symptomatic 6 months or less before diagnosis; annual surveillance of asymptomatic patients may be insufficient. Thus, surveillance by means of regular CT or magnetic resonance imaging (MRI) cannot be advocated. FAP family members who do not yet have polyposis but have signs or symptoms suggestive of a brain tumor should be evaluated with neuroimaging because brain tumors present before polyposis in more than half of FAP patients. Careful evaluation is also important among FAP families in which one member already has a brain tumor because familial clustering occurs. Of such families with FAP-associated brain tumors, 40% had two affected members.[103]
Attenuated Familial Adenomatous Polyposis (AFAP)
Clinical phenotype
AFAP was first described clinically in 1990 in a large kindred with a variable number of adenomas. The average number of adenomas in this kindred was 30, although they ranged in number from a few to hundreds.[205] It has been recommended that the management of AFAP patients include colonoscopy rather than flexible sigmoidoscopy because the adenomas can be predominantly right-sided.[206] Adenomas in AFAP are believed to form around the age of mid-twenties to late twenties.[57] Similar to classic FAP, the risk of CRC is higher in individuals with AFAP; the average age at diagnosis, however, is older than classic FAP at 56 years.[106,107,207] Affected family members have developed CRCs with very few synchronous polyps.[2] Extracolonic manifestations similar to those in classic FAP also occur in AFAP. These manifestations include upper GI polyps (FGPs, duodenal adenomas, and duodenal adenocarcinoma), osteomas, epidermoid cysts, and desmoid tumors.[57] Because of the specific sites of APC pathogenic variants causing AFAP, these patients typically lack CHRPE lesions.
Genetics of AFAP
AFAP is associated with particular subsets of APC pathogenic variants. Three groups of site-specific APC pathogenic variants causing AFAP have been characterized:[106-109,208,209]
- Pathogenic variants associated with the 5’ end of APC and exon 4 in which patients can manifest 2 to more than 500 adenomas, including the classic FAP phenotype and upper GI polyps. Any pathogenic variant in the first four exons,[106] as there is an internal ribosomal entry site in exon 4 that permits the ribosome to skip premature truncation pathogenic variants.[210]
- Exon 9–associated phenotypes in which patients may have 1 to 150 adenomas but no upper GI manifestations.
- 3’ region pathogenic variants in which patients have very few adenomas (<50).
In the absence of family history of similarly affected relatives, the differential diagnosis may include AFAP (including MAP), Lynch syndrome, CMMRD, germline variants in the DNA polymerase proofreading subunits (POLD1 or POLE), or an otherwise unclassified sporadic or genetic problem. A careful family history may implicate AFAP or Lynch syndrome.
APC testing is an important component of the evaluation of patients suspected of having AFAP.[206] If germline APC pathogenic variant testing is negative in suspected AFAP individuals, genetic testing for MUTYH, POLE, and POLD1 pathogenic variants may be warranted.[117]
Clinical management
Patients found to have an unusually or unacceptably high adenoma count at an age-appropriate colonoscopy pose a differential diagnostic challenge.[211,212] The role for and timing of risk-reducing colectomy in AFAP is controversial.[213]
Table 7 summarizes the clinical practice guidelines from different professional societies regarding surveillance of AFAP.
| Organization | Condition | Screening Method | Screening Frequency | Age Screening Initiated | Comment |
|---|---|---|---|---|---|
| FDA = U.S. Food and Drug Administration; IPAA = ileal pouch–anal anastomosis; IRA = ileorectal anastomosis; NCCN = National Comprehensive Cancer Network. | |||||
| aColonoscopy with polypectomy can adequately remove polyps when individuals have a small adenoma burden, which is defined as fewer than 20 adenomas that do not have advanced histology and are each <1 cm in diameter. | |||||
| Europe Mallorca Group (2008) [214] | AFAP | Colonoscopy | Every 2 y; every 1 y if adenomas are detected | 18–20 y | |
| NCCN (2024) [123] | Personal history of AFAP with small adenoma burdena | High-quality colonoscopy and polypectomy | Every 1–2 y | If patient had colectomy with IRA, endoscopic evaluation every 6–12 mo, depending on the patient's polyp burden | |
| Chemoprevention may be considered in patients with progressive polyp burdens to manage the remaining rectum or pouch postcolectomy; at this time, the FDA has not approved medications for this specific indication; NCCN recommends that patients seek the advice of providers with expertise in FAP/AFAP | |||||
| Personal history of AFAP with adenoma burden that cannot be handled endoscopically | Not applicable | Not applicable | Not applicable | Colectomy with IRA preferred. Consider proctocolectomy with IPAA if patient has dense rectal polyposis | |
| Asymptomatic at-risk family member; familial pathogenic variant known; APC pathogenic variant status positive | High-quality colonoscopy | Every 1–2 y if APC positive | Late teens | If adenomas are found, follow AFAP screening guidelines | |
| Asymptomatic at-risk family member; familial pathogenic variant known; APC pathogenic variant status unknown | High-quality colonoscopy | If genetic testing is not performed, colonoscopy can be done every 2 y; if adenomas are found, follow AFAP screening guidelines; if adenomas are not found on multiple subsequent exams, a prolonged screening interval (>2 y) may be considered | Late teens | Discuss benefits of genetic testing | |
MUTYH-Associated Polyposis (MAP)
MAP is an autosomal recessively inherited polyposis syndrome caused by pathogenic variants in the Mut Y homolog gene. The Mut Y homolog gene, which is known as MUTYH, was initially called MYH, but was subsequently corrected because the myosin heavy chain gene already had that designation. MUTYH is located on chromosome 1p34.3-32.1.[215] The protein encoded by MUTYH is a base excision repair glycosylase, which repairs one of the most common forms of oxidative damage. Over one hundred unique sequence variants of MUTYH have been reported. A founder pathogenic variant with ethnic differentiation is assumed for MUTYH pathogenic variants. In White populations of northern European descent, two major variants, Y179C and G396D (formerly known as Y165C and G382D), account for 70% of biallelic pathogenic variants in MAP patients; 90% of these patients carry at least one of these pathogenic variants.[216] Other causative variants that have been found include P405L (formerly known as P391L) (Netherlands),[217,218] E480X (India),[219] Y104X (Pakistan),[220] 1395delGGA (Italy),[221,222] 1186-1187insGG (Portugal),[223] and p.A359V (Japan and Korea).[224-226]
The MUTYH gene was first linked to polyposis in 2002 in three siblings with multiple colonic adenomas and CRC but no APC pathogenic variant.[112] MAP has a broad clinical spectrum. Most often it resembles the clinical picture of AFAP, but it has been reported in individuals with phenotypic resemblance to classical FAP and Lynch syndrome.[227] MAP patients tend to develop fewer adenomas at a later age than patients with APC pathogenic variants [228,229] but still carry a high risk of CRC (35%–75%).[7,230,231] A 2012 study of colorectal adenoma burden in 7,225 individuals reported a prevalence of biallelic MUTYH pathogenic variants of 4% (95% CI, 3%–5%) among those with 10 to 19 adenomas, 7% (95% CI, 6%–8%) among those with 20 to 99 adenomas, and 7% (95% CI, 6%–8%) among those with 100 to 999 adenomas.[116] This broad clinical presentation results from the MUTYH gene's ability to cause disease in its homozygous or compound heterozygous forms. Based on studies from multiple FAP registries, approximately 7% to 19% of patients with an FAP phenotype and without a detectable APC germline pathogenic variant carry biallelic variants in the MUTYH gene.[7,219,229,232]
Adenomas, serrated adenomas, and hyperplastic polyps can be seen in MAP patients.[233] The CRCs tend to be right-sided and synchronous at presentation and seem to carry a better prognosis than sporadic CRC.[215] Clinical management guidelines for MAP recommend screening with high-quality colonoscopy and polypectomy every 1 to 2 years if fewer than 20 adenomas are found. Colonoscopy begins no later than age 25 to 30 years (although screening at a younger age may be considered based on family history).[123,214,230] Upper endoscopic surveillance begins at age 30 to 35 years.[214] (Refer to Table 8 for more information about available clinical practice guidelines for colon surveillance in MAP patients.) The recommended upper endoscopic surveillance interval can be based on the burden of involvement according to Spigelman criteria.[214] Total colectomy with ileorectal anastomosis or subtotal colectomy may be necessary for patients with MAP depending on overall polyp burden.[230,234]
Although MAP is the only known biallelic (recessive) adenoma cancer predisposition syndrome described to date, there are examples of biallelic cases presenting with childhood tumors in which MMR genes are involved. For more information, see the IHC in constitutional mismatch repair deficiency (CMMRD) syndrome section.
Table 8 summarizes the clinical practice guidelines from different professional societies regarding colon surveillance of biallelic MAP.
| Organization | Condition | Screening Method | Screening Frequency | Age Screening Initiated | Comment |
|---|---|---|---|---|---|
| CRC = colorectal cancer; FDR = first-degree relative; IPAA = ileal pouch–anal anastomosis; IRA = ileorectal anastomosis; NCCN = National Comprehensive Cancer Network. | |||||
| aColonoscopy with polypectomy can adequately remove polyps when individuals have a small adenoma burden, which is defined as fewer than 20 adenomas that do not have advanced histology and are each <1 cm in diameter. | |||||
| Nieuwenhuis et al. (2012) [230] | One MUTYH pathogenic variant (monoallelic/MUTYH heterozygote) | Colonoscopy | Every 1–2 y | ||
| NCCN (2024) [123] | Biallelic MUTYH pathogenic variants; personal history of MAP, small adenoma burdena | High-quality colonoscopy and polypectomy | Every 1–2 y | No later than age 25 to 30 y | If patient had colectomy with IRA, endoscopic evaluation every 6–12 mo, depending on the patient's polyp burden |
| Chemoprevention may be considered in certain individuals (especially those with a high polyp burden postcolectomy), but data are limited in patients with MAP; consider referring patients to a center that has experience with MAP to discuss chemoprevention and surgery options | |||||
| Biallelic MUTYH pathogenic variants; personal history of MAP with adenoma burden that cannot be managed endoscopically | Not applicable | Not applicable | Not applicable | Colectomy with IRA. Consider proctocolectomy with IPAA if patient has dense rectal polyposis. If patient had colectomy with IRA, endoscopic evaluation of the rectum may be done every 6–12 mo based on polyp burden | |
| Asymptomatic, at-risk family member; familial pathogenic variant known; MUTYH pathogenic variant status unknown | High-quality colonoscopy | Every 1–2 y | No later than age 25–30 y | Repeat screening every 1–2 years if polyps are not found; the screening interval can be lengthened if an individual does not have polyps on multiple subsequent colonoscopies, based on a provider's judgment; if polyps are found, use MAP screening guidelines. Discuss benefits of genetic testing if the patient's pathogenic variant status is unknown | |
| Asymptomatic, at-risk family member; familial pathogenic variant known; no MUTYH pathogenic variant found | General population screening | ||||
| One MUTYH pathogenic variant (monoallelic/MUTYH heterozygote); proband has a personal history or an FDR with CRC/colon polyps | Increased frequency of screening based on the patient's personal or family history of CRC/colon polyps (refer to NCCN guidelines for CRC screening) [235] | ||||
| One MUTYH pathogenic variant (monoallelic/MUTYH heterozygote); proband does not have a personal history or an FDR with CRC/colon polyps | General population screening | ||||
Many extracolonic cancers have been reported in patients with MAP including gastric, small intestinal, endometrial, liver, ovarian, bladder, thyroid, and skin cancers (melanoma, squamous epithelial, and basal cell carcinomas).[236,237] Additionally, noncancerous extracolonic manifestations have been reported in a few MAP patients including lipomas, congenital hypertrophy of the retinal pigment epithelium, osteomas, and desmoid tumors.[221,229,237,238] Female MAP patients have an increased risk of breast cancer.[239] These extracolonic manifestations seem to occur less frequently in MAP than in FAP, AFAP, or Lynch syndrome.[240,241]
Duodenal polyps in MAP
Similar to FAP, individuals with MAP often develop duodenal adenomas, and are at risk of developing duodenal cancer. Given the relatively recent identification of MAP compared with FAP, the incidence of duodenal polyps and risk of duodenal cancer in MAP is less well defined. Small case series have suggested the incidence of duodenal polyps in MAP to be approximately 30%, considerably lower than that of FAP. In a registry-based study the prevalence of duodenal polyps was 17%; however, only 50% of individuals in this study had undergone an upper GI endoscopy, suggesting the incidence of duodenal polyps was likely underestimated. The lifetime risk of duodenal cancer was estimated to be 4%.[237]
A registry study from the United Kingdom and the Netherlands explored incidence of duodenal polyps and duodenal cancer in a group of patients with MAP who were undergoing regular duodenal surveillance.[242] Of 92 patients, 31 (34%) had evidence of duodenal polyps. The median age at duodenal adenoma detection was 50 years, and in 65% of patients, duodenal adenomas were diagnosed at baseline endoscopy. Eighty-four percent of patients had Spiegelman stage I or stage II polyposis at first detection of polyps, with no patients with stage IV polyposis and no high-grade dysplasia detected. In subsequent surveillance only two patients progressed to Spiegelman stage IV polyposis, after 3.6 and 7.0 years, respectively. There additionally appeared to be sparing of the ampulla, with only two individuals having diminutive polyps without dysplasia in the ampulla. No cancers were detected in patients enrolled in upper GI surveillance programs within these registries. Two individuals with MAP were diagnosed with ampullary and duodenal cancer respectively at ages 83 and 63 years at the time of first-ever upper GI endoscopies. Therefore, duodenal polyps appear less frequently in MAP when compared with FAP; duodenal polyps also appear at a later age in MAP. Based on these results, the authors suggest upper GI endoscopic screening in MAP be initiated at age 35 years.
Because MAP has an autosomal recessive inheritance pattern, siblings of an affected patient have a 25% chance of also carrying biallelic MUTYH pathogenic variants and should be offered genetic testing. Similarly, testing can be offered to the partner of an affected patient so that the risk in their children can be assessed.
The clinical phenotype of monoallelic MUTYH pathogenic variants is less well characterized with respect to incidence and associated clinical phenotypes, and its role in susceptibility to polyposis and colorectal carcinoma remains unclear. Approximately 1% to 2% of the general population carry a pathogenic variant in MUTYH.[7,112,229] A 2011 meta-analysis found that carriers of monoallelic MUTYH pathogenic variants are at modestly increased risk of CRC (odds ratio [OR], 1.15; 95% CI, 0.98–1.36); however, given the rarity of carriers of monoallelic pathogenic variants, they account for only a trivial proportion of all CRC cases.[243] A large study of 2,332 heterozygotes among 9,504 relatives of 264 CRC cases with a MUTYH pathogenic variant found that the risk of CRC at age 70 years was 7.2% for men and 5.6% for women, irrespective of family history. Among those with an FDR with a CRC diagnosis before age 50 years, the risk at age 70 years was 12.5% for men and 10% for women.[231] Caution should be exercised in the interpretation of this study as the vast majority of carrier status from this study was imputed and not based on genotype. The authors felt the risk for MUTYH heterozygotes with an FDR with CRC was sufficiently high to warrant more intensive surveillance than the general population (but the same as for anyone with an FDR with CRC diagnosed before age 50 y).[228,231]
MMR genes may interact with MUTYH and increase the risk of CRC. An association between MUTYH and MSH6 has been reported. Both proteins interact together in base excision repair processes. A study reported a significant increase of MSH6 pathogenic variants in carriers of monoallelic MUTYH pathogenic variants with CRC compared with noncarriers with CRC (11.5% vs. 0%; P = .037).[244] However, a German study failed to duplicate these findings.[245] Additionally, a larger study found no increased cancer risk for carriers of MMR pathogenic variants with a MUTYH variant compared with those with a MMR pathogenic variant alone.[246]
Lynch Syndrome
Introduction
Lynch syndrome is the most common inherited CRC syndrome and accounts for approximately 3% of all newly diagnosed cases of CRC. It is an autosomal dominant condition caused by pathogenic variants in the MMR genes MLH1 (mutL homolog 1), MSH2 (mutS homolog 2), MSH6 (mutS homolog 6), and PMS2 (postmeiotic segregation 2), as well as the gene EPCAM (epithelial cellular adhesion molecule, formerly known as TACSTD1), in which deletions in EPCAM cause epigenetic silencing of MSH2. Lynch syndrome is also associated with a predisposition for developing several extracolonic manifestations, including sebaceous adenomas and cancers of the endometrium and ovaries, stomach, small intestine, transitional cell carcinoma of the ureters and renal pelvis, hepatobiliary system, pancreas, and brain. Lynch syndrome–associated cancers exhibit MSI; therefore, tumor testing is a key component in the diagnosis of Lynch syndrome, in addition to family history. Universal tumor testing of all CRCs is now recommended as a strategy to screen for Lynch syndrome and identify those individuals who may subsequently benefit from germline genetic testing. Intensive cancer screening and surveillance strategies, including frequent colonoscopy, along with risk-reducing surgeries, are mainstays in patients with Lynch syndrome.
History of Lynch syndrome
Between 1913 and 1993, numerous case reports of families with apparent increases in CRC were reported. As series of such reports accumulated, certain characteristic clinical features emerged: early age at onset of CRC; high risk of synchronous (and metachronous) colorectal tumors; preferential involvement of the right colon; improved clinical outcome; and a range of associated extracolonic sites including the endometrium, ovaries, other sites in the GI tract, uroepithelium, brain, and skin (sebaceous tumors). Terms such as cancer family syndrome and hereditary nonpolyposis colorectal cancer (HNPCC) were used to describe this entity.[247]
The term Lynch syndrome replaced HNPCC and is applied to cases in which the genetic basis can be confidently linked to a germline pathogenic variant in a DNA MMR gene. Moreover, HNPCC is misleading as many patients have polyps and many have tumors other than CRC.
With the increased recognition of families that were considered to have a genetic predisposition to the development of CRC, research for a causative etiology led to the development of the Amsterdam criteria in 1990.[248] The Amsterdam criteria were originally used for the identification of high-risk families and included fulfillment of all of the following: three or more cases of CRC over two or more generations, with at least one diagnosed before age 50 years, and no evidence of FAP.
In 1987, a chromosomal deletion of a small segment of 5q led to the detection of a genetic linkage between FAP and this genomic region,[249] from which the APC gene was eventually cloned in 1991.[250] This led to searches for similar linkage in families suspected of having Lynch syndrome who had multiple cases of CRC inherited in an autosomal dominant fashion and young onset of cancer development. The APC gene was one of several genes (along with DCC and MCC) evaluated in families that fulfilled Amsterdam criteria, but no linkage was found among the Lynch kindreds. In 1993, an extended genome-wide search resulted in the recognition of a candidate chromosome 2 susceptibility locus in large families. Once MSH2, the first Lynch syndrome–associated gene, was sequenced, it was evident from the somatic variant patterns in the CRC tumors that the MMR family of genes was likely involved. Additional MMR genes were subsequently linked to Lynch syndrome, including MLH1, MSH6, and PMS2. Lynch syndrome now refers to the genetic disorder caused by a germline variant in one of these DNA MMR genes, distinguishing it from other familial clusters of CRC.
In 2009, a germline deletion in the EPCAM gene was identified as another cause of MSH2 inactivation in the absence of a germline pathogenic variant in MSH2. The variant in EPCAM led to hypermethylation of the MSH2 promoter. Thus, EPCAM, which is not a DNA MMR gene, is also implicated in Lynch syndrome and is now routinely tested in at-risk patients along with the DNA MMR genes listed above.
Defining Lynch syndrome families
Families with a preponderance of CRC and a possible genetic predisposition were initially categorized as having Lynch syndrome based on family history criteria, as well as personal history of young-onset CRC. With the advent of molecular tumor diagnostic testing and the discovery of the germline alterations associated with Lynch syndrome, the clinical criteria have currently fallen out of favor due to their underperformance. However, their use, or the risk estimates provided by the Lynch syndrome prediction models, may be applicable among individuals without personal history of cancer but with a family history suggestive of Lynch syndrome, or for those individuals with CRC but without available tumor for molecular diagnostic testing. (Refer to the Universal tumor testing to screen for Lynch syndrome and the Clinical risk assessment models that predict the likelihood of an MMR gene pathogenic variant sections of this summary for more information.)
The first criteria for defining Lynch syndrome families were established by the International Collaborative Group meeting in Amsterdam in 1990 and are known as the Amsterdam criteria.[248] These research criteria were limited to diagnoses of familial CRC. In 1999, the Amsterdam criteria were revised to include some extracolonic cancers, predominantly endometrial cancer.[251] These criteria provide a general approach to identifying Lynch syndrome families, but they are not considered comprehensive; nearly half of families meeting the Amsterdam criteria do not have detectable pathogenic variants.[252]
Amsterdam criteria I (1990):
- One family member diagnosed with CRC before age 50 years.
- Two affected generations.
- Three affected relatives, one of them an FDR of the other two.
- FAP should be excluded.
- Tumors should be verified by pathological examination.
Amsterdam criteria II (1999):
- Same as Amsterdam criteria I, but tumors of the endometrium, small bowel, ureter, or renal pelvis can be used to substitute an otherwise qualifying CRC.
These criteria were subsequently used beyond research purposes to identify potential candidates for microsatellite and germline testing. However, the Amsterdam criteria failed to identify a substantial proportion of Lynch syndrome kindreds; families that fulfilled Amsterdam criteria I but did not have evidence of MSI and were without a pathogenic germline variant in a DNA MMR gene, were referred to as familial colorectal cancer type X (FCCX).
With the hallmark feature of MSI associated with Lynch syndrome tumors, and the limitations of the Amsterdam criteria related to low sensitivity, the Bethesda guidelines were introduced in 1997. The Bethesda guidelines are a combination of clinical, histopathologic, and family cancer history features that identify cases of CRC that warrant MSI tumor screening. The Bethesda guidelines (with a subsequent revision in 2004) were formulated to target patients in whom evaluation of CRC tumors for MMR deficiency should be considered, and to improve the sensitivity of clinical criteria used to identify individuals who are candidates for mutational DNA analysis.[253,254] (Refer to the Genetic and molecular testing for Lynch syndrome section of this summary for more information about testing for MSI and IHC.)
Bethesda guidelines (1997):
- Cancer in families that meet the Amsterdam criteria.
- The presence of two Lynch syndrome–related cancers, including synchronous and metachronous CRCs or associated extracolonic cancers. [Note: Endometrial, ovarian, gastric, hepatobiliary, or small-bowel cancer or transitional cell carcinoma of the renal pelvis or ureter.]
- The presence of CRC and an FDR with CRC and/or Lynch syndrome–related extracolonic cancer and/or a colorectal adenoma; one of the cancers diagnosed before age 45 years, and the adenoma diagnosed before age 40 years.
- CRC or endometrial cancer diagnosed before age 45 years.
- Right-sided CRC with an undifferentiated pattern (solid/cribriform) on histopathology diagnosed before age 45 years. [Note: Solid/cribriform defined as poorly differentiated or undifferentiated carcinoma composed of irregular, solid sheets of large eosinophilic cells and containing small gland-like spaces.]
- Signet-ring–cell CRC diagnosed before age 45 years. [Note: Composed of more than 50% signet ring cells.]
- Adenomas diagnosed before age 40 years.
Revised Bethesda guidelines (2004)*:
- CRC diagnosed in an individual younger than 50 years.
- Presence of synchronous, metachronous colorectal, or other Lynch syndrome–associated tumors.**
- CRC with MSI-high (MSI-H) pathological associated features diagnosed in an individual younger than 60 years. [Note: Presence of tumor-infiltrating lymphocytes, Crohn-like lymphocytic reaction, mucinous/signet-ring differentiation, or medullary growth pattern.]
- CRC or Lynch syndrome–associated tumor** diagnosed in at least one FDR younger than 50 years.
- CRC or Lynch syndrome–associated tumor** diagnosed at any age in two FDRs or second-degree relatives (SDRs).
*One criterion must be met for the tumor to be considered for MSI testing.
**Lynch syndrome–associated tumors include colorectal, endometrial, stomach, ovarian, pancreatic, ureter and renal pelvis, biliary tract, and brain tumors; sebaceous gland adenomas and keratoacanthomas in Muir-Torre syndrome; and carcinoma of the small bowel.[254,255]
Although the Bethesda guidelines were able to identify a higher proportion of Lynch syndrome carriers than the Amsterdam criteria, they still missed approximately 30% of Lynch syndrome families.[256] Furthermore, the Bethesda guidelines were not consistently used in clinical practice to identify the subset of individuals with CRC who should have MSI tumor testing; the guidelines were deemed cumbersome and difficult to remember by health care providers and the opportunity to refer for genetic evaluation was missed.[257]
With the advent of alternative approaches, including universal testing of all newly diagnosed cases of CRC for MSI (regardless of age at diagnosis or family history of cancer), clinical criteria for Lynch syndrome have been rendered obsolete. While the Bethesda guidelines were intended for individuals with cancer, their performance in individuals unaffected by cancer may still be of use. Given the limited modalities available to assess unaffected individuals for Lynch syndrome, family history and the use of clinical criteria may be appropriate in identifying those who warrant further genetic evaluation and testing.
Clinical risk assessment models that predict the likelihood of an MMR gene pathogenic variant
Because health care providers ineffectively use clinical criteria to select individuals with CRC for genetic referral and evaluation for Lynch syndrome, computer-based clinical prediction models were developed and introduced in 2006 as alternative modalities to provide systematic genetic risk assessment for Lynch syndrome. The risk models include the PREMM (PREdiction Model for gene Mutations) models, MMRpredict, and MMRpro.[258-261]
Four models (PREMM[1,2,6], PREMMplus, MMRpredict, and MMRpro) quantify an individual’s probability of carrying a pathogenic variant in one of the following MMR genes: MLH1, MSH2, and MSH6. The PREMM(1,2,6) model was subsequently extended to include prediction of pathogenic PMS2 and EPCAM variants (PREMM5).[261] While PREMM5, MMRpredict, and MMRpro are specific to Lynch syndrome, PREMMplus quantifies an individual’s risk of having a pathogenic variant in one of 19 genes (including all five Lynch syndrome genes).[262] However, it is unclear if one of these models is preferred for predicting when an individual has Lynch syndrome.
While the models were all created for the same purpose, they differ in the way they were developed and the variables used to predict risk. In addition, the populations in which they were validated reveal each model’s specific characteristics that may impact accuracy.[262-272] Deciding on which model to use in the risk assessment process depends on both the clinical setting in which it is applied and the patient population that is being evaluated. MMRpro’s predictions account for family size and unaffected relatives, the possibility of including molecular tumor data in the risk analysis, and the option of predicting pathogenic variant carrier status following germline testing. The major limitation in the widespread use of MMRpro in routine practice is the need to input data from the entire pedigree (including individuals without cancer), which is relatively time-consuming. Its best use is likely to be as a genetic counseling tool in a specialized high-risk clinic or research setting, as its accessibility is also limited. PREMM’s major advantages include that it is easy to use, available as an online tool, and has been extensively validated, including in a self-administered setting in a GI clinic.[273] It includes risk prediction based on personal and family cancer history up to SDRs for a broad spectrum of extracolonic cancers. However, the model does not consider family size and may overestimate the likelihood of a pathogenic variant in a pedigree that includes multiple elderly family members who are unaffected by CRC or endometrial cancer. Given the ease with which one can use the PREMM model (it has been deemed less time-consuming than MMRpro in validation studies),[268] it may be used by diverse health care providers whose primary aim is to identify patients who should be referred for genetic evaluation, and is likely to be most useful in the pretesting decision-making process. MMRpredict’s use may be limited overall because of its less accurate risk estimates [274] when used to evaluate families with Lynch syndrome–associated cancers and older individuals affected by CRC; the model was developed using data from young-onset CRC cases (patients diagnosed at age <55 y) and did not include extracolonic malignancies. Furthermore, the model does not incorporate tumor testing results or provide post-hoc risk estimates based on gene sequencing results. Lastly, PREMMplus assesses the likelihood of finding a pathogenic variant on a multigene panel of 19 high- and moderate-penetrance genes, and it is not limited to genes associated with CRC.[262]
Overall, there is ample evidence that each of the models has superior performance characteristics of sensitivity, specificity, and positive and negative predictive values that support their use when compared with the existing clinical guidelines for diagnosis and evaluation of Lynch syndrome. Because of the diverse clinical settings in which a health care provider could assess an individual for Lynch syndrome, prediction models offer a potentially feasible and useful strategy to systematically identify at-risk individuals, whether or not they are affected with CRC.
Summary
In conclusion, the presence of tumor MSI in CRCs, along with a compelling personal and family history of cancer, warrants germline genetic testing for Lynch syndrome, and most clinical practice guidelines provide for such an approach. These guidelines combine genetic counseling and testing strategies with clinical screening and treatment measures. Providers and patients alike can use these guidelines to better understand available options and key decisions. (Refer to Table 13 for more information about practice guidelines for diagnosis and colon surveillance in Lynch syndrome.)
Genetics of Lynch syndrome
The genetics of both the tumor and the germline have an important role in the development and diagnosis of Lynch syndrome. Tumor DNA in Lynch syndrome–associated tumors exhibits characteristic MSI, and in these cases, there is typically loss of IHC expression for one or more of the proteins associated with the MMR genes. Molecular testing with MSI and/or IHC has been adopted as a universal screen for diagnosis of Lynch syndrome in newly diagnosed patients with CRC and endometrial cancer. IHC testing results can potentially direct gene-specific germline testing. Many genetic testing laboratories offer multigene (panel) tests that simultaneously test for pathogenic variants in all of the Lynch syndrome–associated genes (and often additional genes associated with inherited cancer susceptibility).
Genetic and molecular testing for Lynch syndrome
MSI
The presence of MSI in colorectal tumor specimens is a hallmark feature of Lynch syndrome and can be cause for suspicion of a germline pathogenic MMR gene variant. Microsatellites are short, repetitive sequences of DNA (mononucleotides, dinucleotides, trinucleotides, or tetranucleotides) located throughout the genome, primarily in intronic or intergenic sequences.[275,276] The term MSI is used when colorectal, endometrial, or metastatic tumor DNA [277] shows insertions or deletions in microsatellite regions when compared with normal tissue. MSI indicates probable defects in MMR genes, which may be due to somatic variants, germline variants, or epigenetic alterations.[278] In most instances, MSI is associated with absence of protein expression of one or more of the MMR proteins (MSH2, MLH1, MSH6, and PMS2). However, loss of protein expression may not be seen in all tumors with MSI and not all tumors with loss of protein expression on IHC will be microsatellite unstable.
Certain histopathologic features are strongly suggestive of MSI phenotype, including the presence of tumor-infiltrating lymphocytes (refer to Figure 4), Crohn-like reaction, mucinous histology, and histological heterogeneity.[279]
Initial designation of a colorectal adenocarcinoma as microsatellite unstable was based on the detection of a specified percentage of unstable loci from a panel of three dinucleotide and two mononucleotide repeats that were selected at a National Institutes of Health (NIH) Consensus Conference and referred to as the Bethesda panel. If more than 30% of a tumor's markers were unstable, it was scored as MSI-H; if at least one, but fewer than 30% of markers were unstable, the tumor was designated MSI-low (MSI-L). If no loci were unstable, the tumor was designated microsatellite stable (MSS). Most tumors arising in the setting of Lynch syndrome will be MSI-H.[280] The clinical relevance of MSI-L tumors remains controversial; the probability is very small that these tumors are associated with a germline pathogenic variant in an MMR gene.
The original Bethesda panel has been replaced by a pentaplex panel of five mononucleotide repeats,[280] which has improved the detection of MSI-H tumors.
(Refer to the Prognostic and therapeutic implications of MSI section of this summary for more information about the treatment implications of MSI testing.)
(Refer to the Universal tumor testing to screen for Lynch syndrome section of this summary for information about the utilization of MSI status in the diagnostic workup of a patient with suspected Lynch syndrome.)
IHC
IHC methods are cheaper, easier to understand, and more widely available as a surrogate for MSI and, for these reasons, have replaced polymerase chain reaction (PCR)–based MSI testing in most institutions. IHC is performed in the colorectal or endometrial tumor (or metastatic sites) [277] for protein expression using monoclonal antibodies for the MLH1, MSH2, MSH6, and PMS2 proteins. Isolated loss of expression of any one of these proteins may suggest which specific MMR gene is altered in a particular patient.[281-284] However, certain proteins can form heterodimers (or have other binding partners) and yield loss of two proteins expressed on IHC.
MSI can lead to nucleotide-pairing slippage (looping) in which single nucleotide mispairs are introduced. Heterodimers of MMR proteins are formed to identify the errors and bind the DNA at these sites.[278,285] For example, MSH2 protein complexes with MSH6 protein to form MutSα, which has the main ability to repair single base pair mismatches and single base pair loop-out lesions that can occur during the replication of a mononucleotide repeat sequence. In the absence of MSH6 protein, the MSH2 protein will dimerize with the MSH3 protein forming the MutSβ complex, which can trigger repair of larger loop-out DNA mismatches, but also has some overlapping activity to repair lesions usually repaired by MutSα.
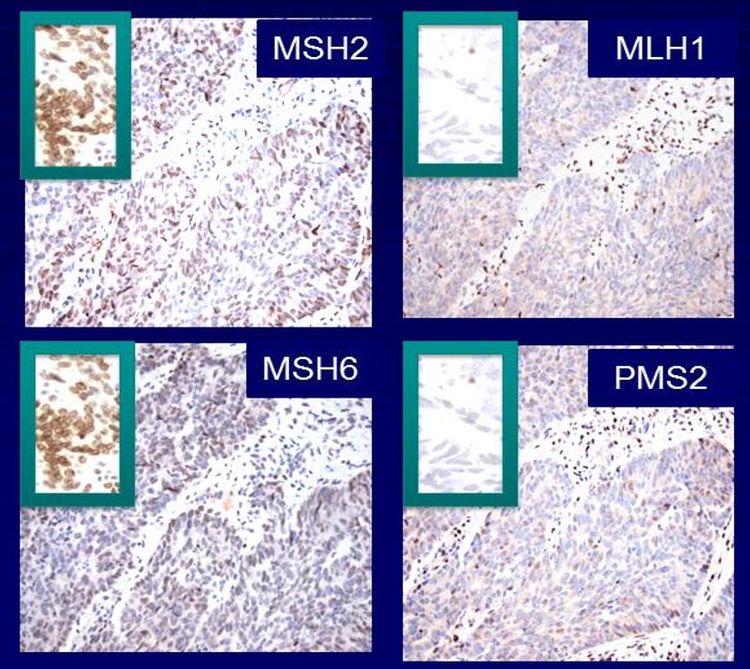
As a result, when the germline pathogenic variant is in the MSH2 gene, the tumor IHC may not express both MSH2 and MSH6, as the latter protein requires binding to MSH2 for stability. In this case, if no pathogenic variant is found in either gene, germline pathogenic variant testing for EPCAM should be considered if it was not already included. Approximately 20% of patients with absence of MSH2 and MSH6 protein expression by IHC and no MSH2 or MSH6 pathogenic variant identified will have germline deletions in EPCAM.[286] The latter mechanism accounts for approximately 5% of all Lynch syndrome cases.[286] A deletion of one exon 9 allele in the EPCAM (TACSTD1) gene, which is immediately upstream of MSH2's start site and in the same orientation, can lead to transcriptional read-through and methylation of the MSH2 promoter. This can lead to subsequent silencing of MSH2 in any tissue that expresses EPCAM. The presence of EPCAM pathogenic variants showing similar methylation-mediated MSH2 loss has been reported in numerous families.[287] On the strength of these observations, germline EPCAM testing is performed in patients with loss of MSH2 protein expression on IHC testing of their CRCs but who lack a detectable MSH2 germline pathogenic variant and is included with MSH2 testing in all colon cancer gene panels. For more information, see the EPCAM section.
In patients with no variants in any of these genes, tumor sequencing may reveal biallelic, somatic MSH2 variants. For more information, see the Somatic biallelic mismatch repair deficiency (sometimes called Lynch-like syndrome) section.
Similarly, the loss of MLH1 (either by germline pathogenic variant or hypermethylation of the MLH1 promoter) results in the absence of expression of both MLH1 and PMS2 proteins in the tumor. The most common abnormal IHC pattern for DNA MMR proteins in colorectal adenocarcinomas is loss of expression of MLH1 and PMS2. PMS2 and MLH1 function as a stable heterodimer known as MutLα. MutLα binds to MutSβ and guides excision repair of the newly synthesized DNA strand.[278] A functional defect in MLH1 results in degradation of both MLH1 and PMS2, while a defect in PMS2 negatively affects only PMS2 expression. Thus, a loss of MLH1 and PMS2 indicates that there is an alteration in MLH1 (promoter hypermethylation or germline variant), while loss of PMS2 expression often indicates that there is a germline PMS2 variant. However, among 88 individuals with PMS2-deficient CRC, PMS2 germline pathogenic variant testing followed by MLH1 germline pathogenic variant testing revealed pathogenic PMS2 variants in 49 individuals (74%) and MLH1 pathogenic variants in 8 individuals (12%).[288] Eighty-three percent of the alterations in MLH1 were missense variants, but two relatives carried identical MLH1 variants, and one individual, who developed two tumors with retained MLH1 expression, carried an intronic variant that led to skipping of exon 8.[288] Therefore, in CRCs with solitary loss of PMS2 expression, an MLH1 germline pathogenic variant should be sought if no PMS2 germline variant is found. Tumors with MSI and loss of MSH2 and MSH6 protein expression are generally indicative of an underlying MSH2 germline variant (inferred MSH2 pathogenic variant). Unlike the case with MLH1, MSI with MSH2 loss is rarely associated with somatic hypermethylation of the promoter.
Unlike MLH1 and MSH2 (which both dimerize with other proteins or have other binding partners), germline pathogenic variants in MSH6 and PMS2 result in the isolated loss of those specific proteins by IHC. However, tumors from MSH6 pathogenic variant carriers may not display the MSI phenotype at a frequency as high as MLH1 and MSH2 carriers (despite an inactive DNA MMR system), as there are pathogenic missense variants that do not completely abrogate protein expression yielding false negative results by IHC testing.[267,289] In a study that reported tumor testing results among MMR germline carriers enrolled through the Colon Cancer Family Registry, 7 of 24 carriers (28%) with MSH6 pathogenic variants had tumors that displayed normal protein expression on IHC staining. IHC tumor testing was more informative for MLH1 and MSH2 pathogenic variant carriers in which 93% of MLH1 carriers had correlating loss of MLH1 protein expression and 96% of MSH2 carriers had loss of MSH2 protein expression.[267]
In some cases, tumors manifest MSI and/or IHC shows loss of DNA MMR protein expression, but no germline pathogenic variant is identified. This tumor phenotype is predominantly due to biallelic somatic inactivation of the DNA MMR genes and is not a pathogenic germline alteration. This phenomenon has been labeled Lynch-like syndrome (sometimes called LLS in the literature), although this terminology may cause confusion since this term represents a mechanism of sporadic carcinogenesis, rather than a stand-alone genetic syndrome. For more information, see the Somatic biallelic mismatch repair deficiency (sometimes called Lynch-like syndrome) section.
| Loss of Protein Expression | Germline MMR Defect Predicted by IHC Protein Expression Loss | ||||
|---|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | PMS2 | EPCAM | |
| IHC = immunohistochemistry; MMR = mismatch repair. | |||||
| MLH1/PMS2 | X | ||||
| MSH2/MSH6 | X | X | |||
| MSH6 | X | ||||
| PMS2 | X | X | |||
| MLH1 | X | X | |||
| MSH2 | X | X | |||
Somatic MLH1 hypermethylation
It is important to recognize that hypermethylation of the MLH1 promoter, a somatic event confined to the tumor, can lead to abnormal protein expression of MLH1 on IHC. Approximately 10% to 15% of sporadic CRC cases have a microsatellite unstable tumor phenotype due to MLH1 hypermethylation and are not heritable. These sporadic MSI colon cancers [290] have a generalized excess of DNA methylation referred to as CIMP.[291] (Refer to the CIMP and the serrated polyposis pathway section in the Introduction section of this summary for more information.) Because loss of MLH1 protein expression on IHC occurs in both Lynch syndrome and sporadic tumors, its specificity for predicting germline MMR gene variants is lower than for the other MMR proteins, and additional molecular testing is often necessary to clarify the etiology of MLH1 absence.
BRAF pathogenic variants have been detected in 68% of CRC tumors with MLH1 promoter hypermethylation and very rarely, if ever, in CRC from patients with Lynch syndrome.[292-295] This suggests that detection of somatic BRAF V600E variant detection in CRC may be useful in excluding individuals from germline variant testing. As a result, BRAF V600 testing and/or MLH1 hypermethylation assays are increasingly used in universal Lynch syndrome–testing algorithms in an attempt to distinguish between an absence of MLH1 protein expression caused by hypermethylation and germline MLH1 pathogenic variants. Making such a distinction is also a more cost-effective approach in excluding individuals from germline testing.
Somatic biallelic mismatch repair deficiency (sometimes called Lynch-like syndrome)
Prior to the widespread use of somatic tumor sequencing, somatic MLH1 hypermethylation was thought to be the only major sporadic pathway for a tumor to have MSI-H/MMR-deficient (MMR-D) biology. It was presumed that any MSI-H/MMR-D CRC or endometrial cancer with patterns of MMR deficiency (other than MLH1/PMS2 loss) was due to Lynch syndrome, even if a germline pathogenic variant in an MMR gene could not be identified. However, as IHC is more routinely performed for all CRCs and endometrial cancers, it has become clear that other patterns of MMR deficiency can arise via sporadic mechanisms, typically by somatic biallelic MMR gene inactivation.[296] This phenomenon has been labeled Lynch-like syndrome (sometimes called LLS in the literature), although this terminology may cause confusion since this term represents a mechanism of sporadic carcinogenesis, rather than a stand-alone genetic syndrome.
Work from the Ohio Colorectal Cancer Prevention Initiative found that 88.4% of CRCs and 100% of endometrial cancers with unexplained MMR-D statuses (i.e., cancers did not have a germline pathogenic variant in an MMR gene or MLH1 promoter hypermethylation) had somatic biallelic MMR deficiency found on somatic tumor sequencing.[297,298] In an additional series of MMR-D cancers (mostly CRCs and endometrial cancers) that underwent paired germline and somatic MMR variant analyses, somatic biallelic genetic alterations were found in 49% of the tumors that lacked causal germline pathogenic variants, MLH1 promoter hypermethylation, and BRAF V600E variants.[299]
When CRCs, endometrial cancers, or other tumors have unexplained MMR-deficiency, in which somatic sequencing fails to identify somatic biallelic MMR deficiency, it is possible that a small fraction of these cases could represent occult Lynch syndrome (or other forms of unidentifiable inherited cancer risk). In these cases, it is recommended that patients' clinical and family histories be carefully examined for possible inherited cancer risk. This will help determine if patients and their families will be clinically managed as if they have suspected Lynch syndrome. In these cases, careful scrutiny of any germline MMR gene VUS is also warranted to assess whether these VUSs could harbor pathogenicity. For more information about VUS genetic test results, see the Value of testing an affected family member first section in Cancer Genetics Risk Assessment and Counseling.
Some studies have attempted to define the risk of CRC and other Lynch syndrome–associated malignancies in cohorts of individuals with Lynch-like syndrome. However, the ability to clinically interpret these data may be limited since these populations presumably include a mix of individuals with sporadic MMR-D cancers and others with inherited cancer risk.[300,301]
IHC in constitutional mismatch repair deficiency (CMMRD) syndrome
In contrast with Lynch syndrome (which is defined by the presence of a single, germline, heterozygous, deleterious variant in an MMR gene), rarely, individuals can have deleterious germline variants in both alleles of the same MMR gene. These cases can present with homozygous or compound heterozygous genotypes. This is termed biallelic mismatch repair deficiency (BMMRD) or constitutional mismatch repair deficiency (CMMRD). The likelihood of CMMRD involving homozygous MMR gene pathogenic variants will inevitably be higher among consanguineous unions. Rates of consanguinity may be higher in rural and geographically and/or culturally isolated populations.[302]
Tumor studies yield characteristic abnormalities. In a series of 28 patients with CMMRD,[115] 17 brain tumors showed loss of staining for MMR protein in normal stromal cells and in neoplastic cells. This differs from tumors in patients with Lynch syndrome, in which normal staining is retained in nontumor cells. In contrast to this characteristic feature seen with IHC, PCR-based MSI analysis was not reliable, as 20 of 28 tumors from patients with CMMRD were MSS. Essentially all of the MSI-H tumors were colon cancers.
The PMS2 gene is markedly overrepresented in cases of CMMRD. It has been suggested that the presence of homozygosity in other MMR gene variants is a prenatally lethal state, while milder expression of PMS2 variants is consistent with survival when present in both parental alleles.
For more information about CMMRD's clinical phenotype, see the CMMRD section.
| Clinical Phenotype | Pathogenic Germline Variant in DNA MMR | Somatic Inactivation of DNA MMR | Tumor Phenotype |
|---|---|---|---|
| CMMRD = constitutional mismatch repair deficiency; FCCX = familial colorectal cancer type X; MMR = mismatch repair; MSI = microsatellite instability; MSS = microsatellite stable. | |||
| aAdapted from Carethers et al.[301] | |||
| Lynch syndrome | Present in one allele | Present in one allele | MSI |
| Sporadic CRC with hypermethylation of MLH1 promoter | Absent | +BRAF | MSI |
| CMMRD | Present in two alleles | Absent | MSI (tumor and normal tissue) |
| Somatic biallelic MMR deficiency (sometimes called Lynch-like syndrome) | Absent | Present in two alleles | MSI |
| FCCX | Absent | Absent | MSS |
Constitutional epimutation
While somatic hypermethylation of the MLH1 promoter is acquired and not uncommon, examples of MLH1 promoter hypermethylation have been described in the germline and are generally not associated with a stable Mendelian inheritance. This constitutional methylation of MMR genes occurs most often in MLH1 and, to a lesser extent, MSH2 and is termed constitutional epimutation.[303] A constitutional epimutation (also referred to as a primary epimutation) is an acquired alteration in normal tissue that silences an active gene or activates an inactive gene.[304] Such epimutations occur most often in maternal alleles. In some cases, all somatic cells appear involved, while in others there is evidence of mosaicism. Tumors in patients with primary epimutations are generally indistinguishable from those otherwise typical of Lynch syndrome germline variant carriers, including age at onset, tumor spectrum, and presence of abnormal MSI and IHC. Since these are not inherited in a Mendelian fashion, antecedent family history of tumors is minimal, and risk to offspring somewhat unpredictable. Epimutations present in a de novo case seem to typically be "erased" in the process of gametogenesis and to not be passed to the next generation. Very rare cases of inherited MLH1 epimutations have been reported.[305,306]
Interpreting molecular alterations in tumors and distinguishing the likely primary epimutation cases from those of sporadic MSI poses significant challenges. Most instances of absence of MLH1 expression are caused by the sporadic hypermethylation of the MLH1 promoter. Rare instances of a de novo constitutional epimutation in MLH1 [307] or an inherited germline MLH1 methylation [308] add some complexity to the interpretation of MSI associated with absence of MLH1 expression. Akin to sporadic MSI, primary epimutation tumors show methylation of the MLH1 promotor and may show BRAF variants as well. As noted above, family history of cancer in such cases tends to be minimal or absent, as in true sporadic MSI. Distinguishing such cases from sporadic cases may call for assaying normal tissue (e.g., blood or normal colon mucosa) for evidence of MLH1 methylation, which will be absent from true sporadic cases and absent from carriers of conventional Lynch syndrome MMR pathogenic variants.
Such MLH1-predominant primary epimutations are to be distinguished from secondary epimutations such as those occurring when MSH2 is methylated as a consequence of inherited variants in the upstream EPCAM gene. (Refer to the EPCAM section of this summary for more information.)
Molecular diagnostic tumor testing to screen for Lynch syndrome in clinical practice
While many molecular pathology laboratories can assess both MSI and IHC, an approach that uses IHC testing as the initial screen for defective MMR activity has been favored because it is less labor intensive and more cost-effective.[309,310] Part of this rationale is that the information provided by IHC may target germline genetic testing toward one specific MMR gene (with the exception of loss of MLH1 expression) as opposed to a comprehensive testing strategy of all Lynch syndrome–related MMR genes that would be directed by the use of MSI alone.[256,309,311-314] While MSI testing was originally favored in the oncologic evaluation of individuals with CRC for its prognostic and therapeutic implications, screening for Lynch syndrome can be more effectively directed by IHC testing.
Universal tumor testing to screen for Lynch syndrome
Use of MSI and/or IHC testing in all newly diagnosed cases of CRC, regardless of the age at diagnosis or family history of cancer, increases the sensitivity of the initial screen for Lynch syndrome. This approach is more sensitive than existing clinical criteria, as many individuals with Lynch syndrome are diagnosed at older ages (>50 y) and have less striking family histories of CRC than previously appreciated. This universal testing of colorectal (and endometrial) tumors using either MSI or IHC testing has been recommended by many professional organizations and is being widely adopted.[123,315-318]
Genetic risk assessment and MMR gene variant testing in individuals with newly diagnosed CRC can lead to improved outcomes for the patient and at-risk family members. Dating back to 2009, the Evaluation of Genomic Applications in Practice and Prevention (EGAPP), a project developed by the Office of Public Health Genomics at the Centers for Disease Control and Prevention (CDC), reported that there was sufficient evidence to recommend offering tumor screening for Lynch syndrome to individuals with newly diagnosed CRC to reduce morbidity and mortality in relatives.[319,320] At that time, there was insufficient evidence to recommend a specific testing strategy between MSI and IHC.
Several studies have demonstrated the feasibility of universal screening for Lynch syndrome. Initial experience from one institution found that among 1,566 patients screened using MSI and IHC, 44 patients (2.8%) had Lynch syndrome. For each proband, an average of three additional family members were subsequently diagnosed with Lynch syndrome.[256] A subsequent pooled analysis of 10,206 incident CRC patients tested with MSI/IHC as part of four large studies revealed a pathogenic variant detection rate of 3.1%.[321] This study compared four strategies for tumor testing for the diagnosis of Lynch syndrome: (1) testing all individuals meeting at least one criterion of the Bethesda guidelines; (2) testing all individuals meeting Jerusalem recommendations;[322] (3) testing all individuals with CRC aged 70 years or younger, or older than 70 and meeting at least one criterion of the Bethesda guidelines; and (4) universal testing of all individuals with CRC.[321] Tumor testing with MSI involved panels individualized at each institution and IHC involved testing all four of the DNA MMR genes involved with Lynch syndrome, across all institutions. The strategy of tumor testing in all individuals diagnosed with CRC at age 70 years or younger and testing individuals over age 70 who met one of the revised Bethesda guidelines yielded a sensitivity of 95.1%, a specificity of 95.5%, and a diagnostic yield of 2.1%. This strategy missed 4.9% of Lynch syndrome cases, but 34.8% fewer cases required IHC/MSI testing, and 28.6% fewer cases underwent germline testing than in the universal approach.
The consideration to further stratify the recommendation for molecular tumor testing by age (i.e., 70 y) warrants attention as it influences the cost-effectiveness of universal screening strategy.
Loss of MLH1 and PMS2 due to somatic hypermethylation is not uncommon, and is more frequently detected with increasing age at CRC diagnosis.[323] Therefore, additional molecular tumor testing including BRAF and MLH1 hypermethylation testing is recommended in cases in which there is loss of MLH1 and PMS2 expression on IHC, thereby decreasing the number of individuals referred for unnecessary germline genetic testing. A testing strategy including MLH1 hypermethylation analyses in individuals aged 70 years or younger with CRC who had loss of MLH1 on IHC was shown to be cost-effective in a population-based study of 1,117 individuals.[324]
Screening individuals with CRC for Lynch syndrome is most often performed in a stepwise fashion based on IHC tumor testing results that evaluate protein expression for the four MMR genes related to Lynch syndrome. One proposed strategy is summarized in Figure 6. This framework does not incorporate a germline testing approach that simultaneously evaluates multiple cancer susceptibility genes (multigene [panel] testing), which may be useful in select patient populations. (Refer to the Multigene [panel] testing section of this summary for more information.)
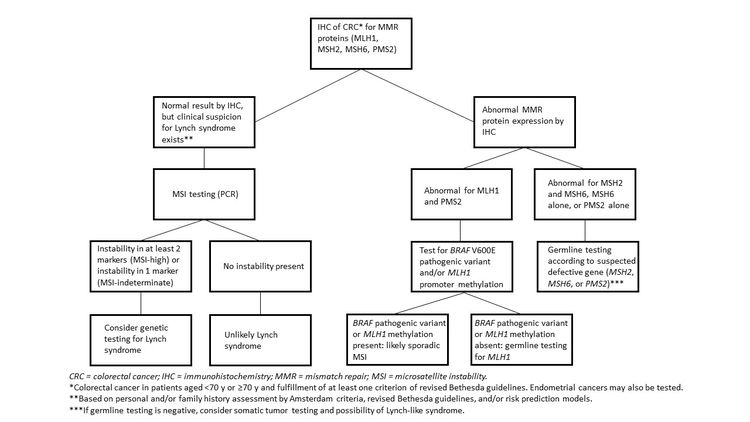
Clinicians are increasingly utilizing tumor sequencing to advance therapeutic decisions in a more personalized approach, particularly in patients with metastatic disease. The performance of next-generation tumor sequencing (NGS) of CRCs for the detection of Lynch syndrome was compared with existing screening protocols that include MSI testing and IHC staining (with BRAF p.V600E testing) in 419 CRC cases recruited in a multicenter, population-based study.[325] Twelve participants were identified as Lynch syndrome carriers by germline DNA testing and all were correctly identified by tumor sequencing, while MSI plus BRAF testing and IHC plus BRAF testing missed five and six Lynch syndrome cases, respectively. Tumor sequencing had a higher sensitivity than IHC plus BRAF testing (100% vs. 89.7%; P = .04) and MSI plus BRAF testing (100% vs. 91.4%; P = .07) while specificity was comparable across all strategies (95.3% for tumor sequencing, 94.6% for IHC plus BRAF, and 94.8% for MSI plus BRAF; P = not significant). In a validation cohort of 46 known Lynch syndrome pathogenic variant carriers with CRC, tumor sequencing yielded similar results and correctly identified 100% of carriers. In addition, the authors highlighted potential therapeutic implications by reporting on somatic alterations identified by tumor sequencing in 283 participants. This study suggested that tumor sequencing is a highly effective mode of identifying Lynch syndrome; however, the cost-effectiveness of this strategy remains to be determined.
A 2019 retrospective study using data from a large, community-based, integrated U.S. health care system compared the diagnostic performance of age-restricted screening strategies for Lynch syndrome by reflex MMR IHC of all CRCs versus a universal screening strategy without an upper age limit.[326] Lynch syndrome identification decreased substantially after age 70 years to age 75 years, with minimal incremental gain after age 80 years. The number of CRCs needed to be screened to identify one Lynch syndrome case was 20 among patients diagnosed with CRC at age 50 years or younger but increased to 208 for those with CRC at age 71 years to age 80 years, and 668 for those diagnosed after age 80 years.
Cost-effectiveness of universal tumor screening for Lynch syndrome
Results are available from a Markov model that incorporated the risks of colorectal, endometrial, and ovarian cancers to estimate the effectiveness and cost-effectiveness of strategies to identify Lynch syndrome among individuals aged 70 years or younger with newly diagnosed CRC.[310] The strategies incorporated in the model were based on clinical criteria, prediction algorithms, and tumor testing or up-front germline pathogenic variant testing followed by directed screening and risk-reducing surgery. IHC followed by BRAF pathogenic variant testing was the preferred strategy in this study. An incremental cost-effectiveness ratio of $36,200 per life-year gained resulted from this strategy. In this model, the number of relatives tested (3–4) per proband was a critical determinant of both effectiveness and cost-effectiveness. These results were similar to earlier analyses conducted by EGAPP which found that the most cost-effective approach was to test all tumors for absence of protein expression of MSH2, MLH1, MSH6, and PMS2 followed by targeted germline testing of MSH2, MLH1, or MSH6 offered depending on which protein was absent. If there was absence of MLH1, testing was offered for BRAF variant-negative tumors.[320]
NCCN 2024 guidelines support using universal screening on all CRCs to help identify individuals who may have Lynch syndrome. Universal screening can include the following testing methods: IHC testing, MSI testing, and comprehensive tumor NGS panel testing.[123] Universal screening in all individuals irrespective of age was associated with a doubling of incremental cost per life-year saved compared with screening only those younger than 70 years.[310] The authors of this analysis conclude that screening individuals younger than 70 years appears reasonable, while screening all individuals regardless of age might also be acceptable, depending on willingness to pay.
However, it is important to note that the conclusions from this study were contingent upon the number of at-risk relatives who underwent germline testing (through a process known as cascade screening) based on the identification of a germline MMR gene variant in the index case of CRC in the family. In their model, to meet the accepted $50,000 cost-effective threshold, testing a minimum of three to four relatives was necessary.[310] This emphasizes the importance of provider-to-patient communication, family communication, and the need to ensure improved uptake of germline testing in Lynch syndrome families with a known causative gene. (Refer to the Psychosocial Issues in Hereditary Colon Cancer Syndromes section of this summary for more information about family communication and uptake of genetic testing in families with Lynch syndrome.)
Another study addressed the cost-effectiveness of testing for pathogenic variants in the Lynch syndrome–associated genes and evaluated 21 screening strategies, including clinical criteria, use of clinical Lynch syndrome prediction models, and molecular tumor testing.[327] The model included two steps: (1) measurement of the newly identified number of Lynch syndrome diagnoses; and (2) measurement of the life-years gained as a result of confirming Lynch syndrome in a healthy carrier. Among all of the strategies modeled, screening the proband with a predictive model such as PREMM(1,2,6) followed by IHC for MMR protein expression and germline genetic testing was the best approach, with an incremental cost-effectiveness ratio of $35,143 per life-year gained. Germline genetic testing on all probands was the most effective approach, but at a cost of $996,878 per life-year gained. The authors concluded that the initial step of Lynch syndrome screening should utilize a predictive model in the proband, and that both universal testing and general population screening strategies were not cost-effective screening strategies for Lynch syndrome.
Establishment of an upper age limit for universal tumor testing remains controversial. Some experts have endorsed testing only individuals with CRC who are younger than 70 years (reserving testing in individuals ≥70 y for only those meeting the revised Bethesda criteria; with this strategy, 5% of carriers would be missed).[328] However, others have advocated against an upper age limit for testing given the potential benefit to younger generations via cascade screening and the opportunity for increased surveillance and other prophylactic interventions in individuals found to carry a known familial pathogenic variant.
Another cost-effectiveness analysis was performed using data from 179 consecutive endometrial cancer patients diagnosed at or before age 70 years and screened with MMR IHC and reflex MLH1 promoter hypermethylation, among whom seven Lynch syndrome carriers (3.9%) were identified.[329] Only one of the seven Lynch syndrome probands was age 50 years or younger at endometrial cancer diagnosis. The authors calculated that screening women diagnosed with endometrial cancer at age 51 to 70 years resulted in an additional 29.3 life-years gained (on top of the 45.4 life-years gained by screening women diagnosed at age ≤50 y), and the incremental cost-effectiveness ratio for screening all diagnoses at age 70 years or younger versus diagnoses at age 50 years or younger was 5,252 euro per life-year gained. Universal tumor-based screening of all women aged 70 years or younger was also cost-effective, compared with strategies using the Bethesda guidelines to guide MMR and MSI testing with an incremental cost-effectiveness ratio of 6,668 euro per life-year gained.
The cost-effectiveness of universal tumor testing in both CRC and endometrial cancer is largely driven by the assumption of cascade screening through which other at-risk family members will be identified, tested, and subsequently pursue their own cancer risk reduction.[310]
The cost of germline genetic testing continues to decrease with advancements in DNA analyses, including simultaneous testing of multiple germline variants associated with malignancy, through multigene (panel) tests. As a result, additional cost-effective analyses using more updated data related to germline testing will need to be conducted. Multigene (panel) testing may become a more favorable and cost-effective approach in the future.
Considerations and limitations related to universal tumor testing for Lynch syndrome
While universal screening continues to be adopted nationally, there is significant variability in the uptake and approach to molecular testing. A 2011 survey of the National Society of Genetic Counselors revealed that more than 25% of respondents had some form of universal screening implemented at their center. Tumor screening methods varied; 34 (64.2%) of 53 centers started with IHC, 11 (20.8%) of 53 centers started with MSI testing, and 8 (15.1%) of 53 centers performed both tests on newly diagnosed colorectal tumors.[330] A 2012 survey suggested that some form of universal screening was being routinely performed at 71% of the National Cancer Institute (NCI) Comprehensive Cancer Centers, but utilization dropped to 15% among a random sample of community hospital cancer programs.[331]
Because adherence to universal screening for Lynch syndrome may be poor (many patients are not referred for genetic evaluation and testing), a prospective quality improvement study utilizing the Six Sigma conceptual framework was conducted to improve the implementation of universal genetic screening among young patients with CRC.[332] The main aim of the study was to increase the proportion of tumor studies for deficient MMR among patients with early-onset CRC (aged 18–50 y). The intervention involved patient and provider education, in addition to visual cues provided at point of care. The study demonstrated an improvement of 21.5% in the rate of IHC testing in young adults with CRC over the 12-month postintervention period compared with the preintervention period.
Studies reporting uptake of genetic testing for Lynch syndrome have largely focused on individuals and families who were selected for potential risk of Lynch syndrome based on family history or clinical characteristics. While universal tumor screening is increasingly being adopted to identify newly diagnosed patients who may have a germline variant, few studies have examined the uptake of genetic testing after universal tumor testing. An important implication of universal screening for Lynch syndrome is that it does not result in automatic germline testing in appropriate individuals. In the clinical setting, more follow-up by health care teams to facilitate referral to genetic counseling for patients with abnormal tumor screening results may improve completion of genetic testing.[333] Higher levels of patient completion of genetic testing after abnormal tumor screening may be associated with having genetic counselors involved in this process to disclose screen-positive results, provide counseling after tumor testing, or facilitate referrals.[334]
Subsequent genetic counseling requires coordination between the pathologist, the referring surgeon or oncologist, and a cancer genetics service. As an illustration, a population-based screening study found that only 54% of patients with an IHC-deficient tumor (that was BRAF pathogenic variant negative) ultimately consented to and proceeded with germline MMR testing.[335] One institution found 21 pathogenic variants among 1,100 patients who underwent routine MSI and IHC testing after a diagnosis of CRC. This study found markedly increased uptake of genetic counseling and germline MMR gene testing when both the surgeon and a genetic counselor received a copy of abnormal MSI/IHC results, especially when the genetic counselor played an active role in patient follow-up.[333]
In contrast to tumor testing, which is commonly performed without a patient's prior knowledge, germline genetic testing, such as germline testing for MMR pathogenic variants, generally includes genetic counseling and requires patient permission before it is performed. A cross-sectional survey of U.S. cancer programs (20 NCI–designated Comprehensive Cancer Centers and 49 community hospital cancer programs) found that, of those that performed MSI and/or IHC testing as part of standard pathological evaluation at the time of colon cancer diagnosis in all or select cases, none required written informed consent before tumor testing.[331]
Diagnostic strategies for all individuals diagnosed with endometrial cancer
Given the increased prevalence of endometrial cancer among carriers of MMR pathogenic variants, there is a growing consensus to screen patients with endometrial cancer for Lynch syndrome.
In a study that examined the feasibility and desirability of performing tumor screening of all endometrial cancers, regardless of age at diagnosis or family history of cancer, at least 2.3% (95% CI, 1.3%–4.0%) of newly diagnosed patients had Lynch syndrome.[336,337] Eight of thirteen cases diagnosed with Lynch syndrome were aged 50 years or older, eight did not meet published family history criteria for Lynch syndrome, and two would have been missed by MSI testing. Because of the increased prevalence of endometrial cancer and the results of this study, the authors support universal screening of endometrial cancers for Lynch syndrome. (Refer to the IHC section of this summary for more information about performing IHC for MMR protein expression.)
Another smaller study of 242 consecutive endometrial cases demonstrated a 4.5% (11/242) prevalence of MMR-deficient cases lacking somatic MLH1 promoter hypermethylation, including four cases (1.7%) with germline MMR variants, four cases (1.7%) with two somatic MMR alterations on NGS, and two cases (0.8%) with otherwise unexplained MMR-deficiency.[338] Such findings demonstrate that universal MMR tumor screening of endometrial cancers will identify individuals with underlying Lynch syndrome and a spectrum of non-Lynch syndrome cases with various forms of MMR-deficiency.
Another study prospectively evaluated universal IHC-based screening of both CRC and endometrial cancer cases, irrespective of age at diagnosis.[339] In both the tertiary and community settings, 1,290 CRC and 484 endometrial cancer cases were screened between 2011 and 2013. The study additionally calculated PREMM(1,2,6) and PREMM5 scores for all patients in whom a germline pathogenic variant was detected. Abnormal staining was observed in 22% of endometrial cancers and 18.8% of CRCs. After excluding those cases felt to be sporadic because of the presence of BRAF and/or hypermethylation of MLH1, 10.8 % of patients with CRC and 6.6% of patients with endometrial cancer were referred for genetic counseling. Lynch syndrome was diagnosed in 24 individuals (1.4%), 66% of whom had CRC. The overall detection rate of Lynch syndrome was 1.7% in endometrial cancer cases and 1.2% in CRC cases. Among Amsterdam criteria, Bethesda guidelines, PREMM(1,2,6), and PREMM5, the best performing model was PREMM5, which would have detected 82% of cases identified by universal screening.
The cost-effectiveness of tumor testing of women diagnosed with endometrial cancer was examined in a model-based simulation study and included IHC testing in the following scenarios: (1) diagnosis before age 50 years; (2) diagnosis before age 60 years; (3) any age at diagnosis with the presence of an FDR with any Lynch syndrome–associated cancer; and (4) all cases irrespective of diagnosis age and family history. Women fulfilling Amsterdam II criteria or those diagnosed before age 50 years with at least one FDR with any Lynch syndrome–associated cancer were directly referred for genetic counseling and genetic testing without IHC testing. A strategy of IHC testing for MMR protein expression in all patients with endometrial cancer and an FDR with any Lynch syndrome–associated cancer was reported to be cost-effective in the detection of Lynch syndrome.[340] This strategy had an incremental cost ratio of $9,126 per life-year gained relative to the least-costly strategy, which was genetic testing on all women diagnosed with endometrial cancer before age 50 years with at least one FDR with a Lynch syndrome–related cancer. Life expectancy was highest with the most inclusive testing strategy of IHC testing of all women with endometrial cancer irrespective of age at diagnosis or family history but had the least favorable incremental cost ratio of $648,494 per life-year gained. NCCN recommends tumor testing with IHC and/or MSI and a comprehensive tumor NGS panel testing for all endometrial cancers.[123] Despite these recommendations, the uptake of universal screening in women newly diagnosed with endometrial cancer is unclear.
(Refer to the PDQ summary on Genetics of Breast and Gynecologic Cancers for more information about endometrial cancer as a component of Lynch syndrome.)
MSI in all cancers
Use of MSI testing across all tumor types has become an important screening tool to select cases that may have a favorable response to immune checkpoint inhibitor therapy. These results may potentially be used to screen for Lynch syndrome in tumors other than CRC. A study evaluated MSI across a wide variety of malignancies and evaluated its use as a potential means to identify Lynch syndrome, regardless of tumor type.[341] In a study of more than 15,000 patients with more than 50 types of cancers evaluated in a single-center study, data on well-annotated tumor and matched normal DNA sequencing results with paired germline MMR gene testing, were used to determine MSI status. MSI was determined using a software tool that reports the percentage of unstable microsatellites as a score from paired tumor-normal genome sequencing data and allows for comprehensive investigation of MSI sites simultaneously. The approach used has been reported to be more sensitive across cancers not typically screened for MMR-deficiency (dMMR) than MSI testing of five mononucleotide microsatellite foci using PCR.[342] CRC and endometrial cancer comprised the majority of cancers with MSI-H in this study, but 38% (125 of 326) of MSI-H tumors and more than 90% of those with intermediate-level MSI were other cancer types. Germline testing confirmed a diagnosis of Lynch syndrome in 16.3% and 1.9% of tumors with MSI-H and intermediate-level MSI, respectively, in addition to 0.3% of cases that lacked MSI. Importantly, half of all Lynch syndrome carriers with MSI-H/intermediate tumors had primary cancers other than CRC or endometrial cancer, with many malignancies not associated with Lynch syndrome. Among those individuals with a noncanonical Lynch syndrome cancer, nearly half failed to meet clinical criteria for Lynch syndrome testing on the basis of their cancer diagnosis or family cancer history. Furthermore, intermediate-level MSI and MSS phenotypes were most often observed in cancers not classically related to Lynch syndrome and in individuals with germline PMS2 variants. This study supports other findings related to the variable phenotypic expression of Lynch syndrome on the basis of the altered MMR gene and its broad constellation of associated malignancies that make it difficult to be identified by clinical criteria alone. In addition, the investigators further analyzed a unique gene variant signature in every tumor and correlated results to the observed MSI phenotype and germline MMR status to provide some indirect data on whether a gene variant carrier’s cancer was caused by Lynch syndrome and MMR deficiency or possibly an incidental finding. This is pertinent in evaluating those cancers whose association with Lynch syndrome is unclear and debatable, such as breast and prostate cancer. The authors’ finding that none of the breast cancer patients with Lynch syndrome in this very large cohort had tumors with MSI lends support to the hypothesis that these individuals’ germline MMR gene variants may simply be incidental findings and not etiologic to their cancer diagnosis.
A single-institution study of 100 histologically confirmed small bowel adenocarcinomas with MSI/MMR tumor studies identified 26 cases with MMR deficiency, 10 of which were due to Lynch syndrome. Most cancers were located in the duodenum, followed by the jejunum and the ileum. The authors recommended that individuals with small bowel adenocarcinomas be screened for Lynch syndrome, given the implications of MMR deficiency on treatment and clinical management.[343]
Germline genetic testing
Genetic testing for germline pathogenic variants in MLH1, MSH2, MSH6, PMS2, and EPCAM can help formulate appropriate intervention strategies for the affected variant-positive individual and at-risk family members, many of whom may be unaffected by cancer.
If a pathogenic variant is identified in an affected person, then testing for that same pathogenic variant should be offered to all at-risk family members. At-risk relatives who test negative for the identified pathogenic variant in the family are not at increased risk of CRC or other Lynch syndrome–associated malignancies and can follow surveillance recommendations applicable to the general population. Family members who carry the familial pathogenic variant are referred to surveillance and management guidelines for Lynch syndrome. (Refer to the Management of Lynch syndrome section of this summary for more information.)
If no pathogenic variant is identified in the affected family member, then testing is considered negative for Lynch syndrome in that individual. With advances made in DNA sequencing technologies, it is unlikely that current gene testing is not sensitive enough to detect a pathogenic variant in the genes tested. Advances in testing, including the common use of NGS by most commercial testing laboratories have improved upon the detection of certain alterations such as large deletions or genomic rearrangements as well as the presence of a pseudogene PMSCL in PMS2.
Possible reasons why a pathogenic variant may not be detected include the following:
- The family could have a variant in a yet-unidentified gene that causes Lynch syndrome or a predisposition to colon cancer.
- The individual tested in the family may have developed colon cancer through a nongenetic mechanism (i.e., it is a sporadic case also known as a phenocopy), while the other cases in the family are really the result of a germline variant. If this scenario is suspected, testing another affected individual who has had a Lynch syndrome–associated cancer is recommended.
- In cases in which a CRC tumor displayed MSI and/or abnormal IHC, but no germline pathogenic variant was detected, biallelic somatic variants may be the resulting etiology. These are sometimes, confusingly, called Lynch-like syndrome cases, even though they are typically not considered to be familial.
Failure to detect a pathogenic variant could mean that the family truly is not at genetic risk despite a clinical presentation that suggests a genetic basis (e.g., the patient may have biallelic somatic variants in an MMR gene). If no variant can be identified in an affected family member, testing should not be offered to at-risk members because results would be uninformative for the relatives. They would remain at increased risk of CRC by virtue of their family history and should continue with recommended intensive screening.
(Refer to the Management of Lynch syndrome section of this summary for more information.)
Multigene (panel) testing
Germline variant analysis of MLH1, MSH2 (including EPCAM), MSH6, and PMS2 may be considered in instances in which tumor tissue is not available from individuals to test for MSI and/or MMR protein IHC. This approach has become less expensive with the advent of multigene (panel) testing, which is now offered by several clinical laboratories at a cost that may be comparable to single-gene testing. The cost of multigene testing may also approach the cost of tumor screening and may prove to be a cost-effective approach in individuals affected by CRC. At present, multigene tests are not routinely recommended for universal screening for Lynch syndrome among all newly diagnosed CRC patients, but they may be very useful in select populations, such as those with early-onset CRC [344] or from familial, high-risk clinic-based populations. It is also important to note that pathogenic variants may be detected in other cancer-associated genes beyond Lynch syndrome. In a study of 1,112 individuals who met NCCN criteria for Lynch syndrome testing and who underwent multigene testing with a 25-gene panel, as expected, 114 individuals (9.0%) were found to have pathogenic variants in MMR genes; however, 71 individuals (5.6%) were found to have a pathogenic variant in non-Lynch syndrome cancer predisposition genes, such as BRCA1, BRCA2, APC, MUTYH (biallelic), and STK11. Lastly, multigene tests yield a high proportion of VUS. In the aforementioned study, a total of 479 patients (38%) had one or more VUS.[345]
Individuals with early-onset CRC have been shown to have a high frequency and wide spectrum of germline pathogenic variants, indicating that panel testing in this population may be beneficial. In a study of 450 patients with early-onset CRC (mean age at diagnosis, 42.5 y) and a family history including at least one FDR with colon, endometrial, breast, ovarian, and/or pancreatic cancer, 75 germline pathogenic or likely pathogenic variants were identified in 72 patients (16%).[344] The spectrum of variants identified included Lynch syndrome and non-Lynch syndrome–associated genes, including several genes that have not traditionally been associated with CRC (e.g., BRCA1/BRCA2, ATM, CHEK2, PALB2, and CDKN2A). Given the high frequency and variety of hereditary cancer syndromes identified, the authors suggested that multigene testing in this population may be warranted. Similarly, another smaller single-institution analysis of 151 individuals with CRC identified pathogenic germline variants in 9.9% of individuals.[346]
Multigene testing has also been examined in a larger study of 1,058 individuals with CRC who were unselected for age at diagnosis, personal or family history, or MSI/MMR test results.[347] Germline pathogenic variants in cancer susceptibility genes were identified in 105 individuals (9.9%). While 33 individuals (3.1%) carried pathogenic variants in Lynch syndrome genes, 74 (7.0%) had pathogenic variants in non-Lynch syndrome–associated genes, including APC, MUTYH, BRCA1/BRCA2, PALB2, CDKN2A, TP53, and CHEK2. These data illustrate the breadth of variants that may be identified in unselected CRC patients; thus, use of a comprehensive multigene test may be warranted.
A 2017 study examined the frequency of pathogenic Lynch syndrome–associated gene variants in individuals undergoing multigene testing at a single commercial United States laboratory between 2012 and 2015, and reported on the characteristics of those carriers identified with Lynch syndrome.[348] The study reports on the largest cohort of individuals tested through multigene testing to date; data was reported on 34,980 individuals who had undergone various multigene panel tests that included the MMR and EPCAM genes, where the indication for testing was not limited to Lynch syndrome. A total of 618 pathogenic variants were identified in 612 individuals (1.7%) and analyses were conducted on 579 subjects (after exclusion of 33 individuals who had a Lynch syndrome–associated variant and a second MMR variant or other pathogenic alteration in another cancer predisposition gene). The majority of carriers were affected by cancer, including non-Lynch syndrome–associated malignancies, where breast cancer was most frequently reported (124/423, 23.5%). MSH6 variants were most prevalent (29.3%), followed by PMS2 (24.2%), MSH2 (23.7%), MLH1 (21.6%), and EPCAM (1.2%). This finding differs from previous data where MSH2 and MLH1 variants were more prevalent, as individuals were more often selected for Lynch syndrome–specific testing due to a personal and/or family history of CRC.
The study reports on genotype-phenotype correlations on 528 Lynch syndrome carriers, the majority of whom had CRC (186, 35.2%) and endometrial cancer (136, 25.8%), followed by breast cancer (124, 23.5%) and ovarian cancer (74, 14%).[348] One hundred forty-five carriers presented with breast or ovarian cancer as their sentinel tumor and did not carry a prior diagnosis of CRC or endometrial cancer prior to the time of multigene testing. When examining MMR gene variant distribution among tumor-specific subgroups, a higher frequency of MSH6 and PMS2 variants were detected in carriers with breast cancer only than MLH1 and MSH2, where the latter pathogenic variants were more frequent in subjects with CRC only. For patients with breast cancer only, the frequency of PMS2 gene variants was significantly higher than population estimates, which was not the case for MLH1, MSH2, or MSH6. A comparable retrospective study reported similar findings. Standardized incidence ratios (SIRs) of breast cancer were calculated by comparing observed breast cancer frequencies in a population of 423 women with pathogenic or likely pathogenic variants in MMR genes with those in the general population. The authors reported a statistically significant age-standardized risk of breast cancer for MSH6 carriers (SIR, 2.11; 95% CI, 1.56–2.86) and PMS2 carriers (SIR, 2.92; 95% CI, 2.17–3.92).[349] A critical limitation of both of these studies was the excess of breast cancer cases in the overall referral population as well as the known high background population prevalence of MSH6 and PMS2 germline pathogenic variants.
Clinical criteria for the identification of Lynch syndrome, including the Amsterdam criteria, revised Bethesda guidelines, or the PREMM(1,2,6) risk prediction model, would have failed to identify 27.3% of Lynch syndrome carriers in this study.[348] Given the increased prevalence of breast and ovarian cancers, 58.9% met the NCCN guidelines for BRCA1/BRCA2 testing and of these, 36.7% also met NCCN guidelines for Lynch syndrome testing. Lastly, there were limited data on tumor testing results, available only on 18.8% of pathogenic variant carriers, where results were often discordant with the altered gene, which was most often reported in MSH6 and PMS2 carriers. Results of this study support the use of multigene testing for Lynch syndrome and further study of the respective cancer risks, as current testing strategies limit identification of Lynch syndrome carriers and associated malignancies.
Lastly, germline MMR genes have been detected unexpectedly among individuals undergoing multigene testing for cancers not commonly associated with Lynch syndrome, such as breast and prostate cancer. As a result, the cancer spectrum associated with Lynch syndrome may be wider than previously appreciated. For more information, see the Breast cancer and Prostate cancer sections and Genetics of Prostate Cancer.
(Refer to the Multigene [panel] testing section in the PDQ summary on Cancer Genetics Risk Assessment and Counseling for more information about multigene testing, including genetic education and counseling considerations, and research examining the use of multigene testing.)
Cost-effectiveness of multigene (panel) testing
As genetic testing becomes routine rather than the exception, questions regarding the cost of testing are inevitable. Historically, a cost-effectiveness ratio of $50,000 per quality-adjusted life-year (QALY) has been used as the benchmark for good value for care.[350] Over time it has been suggested that this threshold is too low and that other thresholds such as $100,000 or $150,000 be used.[350]
A 2015 study evaluated the cost-effectiveness of multigene testing for CRC and polyposis syndromes in patients referred to a cancer genetics clinic.[351] These authors developed a decision model to estimate the immediate and downstream costs for patients referred for evaluation and of CRC surveillance in family members identified as carriers of pathogenic variants. The costs were estimated on the basis of published models from the CDC and from an academic molecular genetics laboratory. They classified the syndromes on the basis of inheritance pattern and penetrance of CRC. Four custom panels were compared with the standard of care. The four panels tested for (1) Lynch syndrome–associated genes only (MLH1, MSH2, MSH6, PMS2, and EPCAM); (2) genes in panel 1 and additional genes associated with autosomal dominant inheritance and high CRC penetrance (APC, BMPR1A, SMAD4, and STK11); (3) genes in panels 1 and 2 and those associated with autosomal recessive inheritance with high CRC penetrance (MUTYH); or (4) all genes in the first three panels and those associated with autosomal dominant conditions with low penetrance (PTEN, TP53, CDH1, GALNT12, POLE, POLD1, GREM1, AKT1, and PIK3CA). The respective costs were as follows: panel 1, $144,235 per QALY; panel 2, $37,467 per QALY; panel 3, $36,500 per QALY; and panel 4, $77,300 per QALY when compared with panel 3. The authors concluded that the use of an NGS multigene test that includes highly penetrant CRC and polyposis syndromes and Lynch syndrome cancer genes was the approach most likely to provide clinically meaningful results in a cost-effective fashion.
The cost of germline genetic testing continues to decrease with advancements in technology since the time this model analysis was conducted; additional studies are needed to continue to assess the cost-effectiveness of this testing approach.
Prevalence, clinical manifestations, and cancer risks associated with Lynch syndrome
Lynch syndrome is an autosomal dominant syndrome characterized by an early age of onset of CRC, excess synchronous and metachronous colorectal neoplasms, right-sided predominance, and extracolonic tumors, notably endometrial cancer. Lynch syndrome is caused by pathogenic variants in the DNA MMR genes, namely MLH1 on chromosome 3p21;[352,353] MSH2 on chromosome 2p22-21;[354,355] MSH6 on chromosome 2p16;[356] and PMS2 on chromosome 7p22.[352-355,357-360] The function of these genes is to maintain the fidelity of DNA during replication. Lynch syndrome is also associated with pathogenic variants of the EPCAM gene on chromosome 2p21, which causes epigenetic silencing of MSH2, located immediately downstream of this gene.[361,362]
Lynch syndrome accounts for about 3% of all newly diagnosed cases of CRC.[309] In earlier studies, the average age of CRC diagnosis in Lynch syndrome pathogenic variant carriers was as young as 44 to 52 years. [256,309,363] In contrast, from 2015 to 2019, the median age of CRC diagnosis was 66 years in individuals with sporadic CRC.[364] In subsequent studies that corrected for ascertainment bias to determine cancer-related risk estimates and genotype-phenotype correlations, the average age at CRC diagnosis was 61 years among Lynch syndrome–associated pathogenic variant carriers.[365]
Original reports related to overall and gene-specific prevalence estimates in Lynch syndrome relied heavily on retrospective data from familial cancer registries worldwide. Earlier risk estimates of CRC (and endometrial cancer) reported in Lynch syndrome were subject to ascertainment bias and overestimation, given that data were derived largely from familial cancer registries and cases were often ascertained based on young-onset CRC or an increased number of CRC cases among relatives. Correction of these cancer risk estimates has been made possible through modified segregation analyses, where statistical methodology provides more accurate estimates and adjusts for ascertainment bias. Conversely, risk estimates related to extracolonic malignancies, with the exception of endometrial cancer, may be prone to underestimation because many families may have underreported these cancers in relatives, and Lynch syndrome–related tumors may have occurred later in life.
In a large population-based study of 5,744 CRC cases who were recruited irrespective of family cancer history from the United States, Australia, and Canada, it was estimated that 1 in 279 individuals in the population carry an MMR pathogenic variant associated with Lynch syndrome.[366]
In another population-based study of 450 individuals with CRC but limited to young onset with diagnoses occurring before age 50 years, germline pathogenic variants were identified in 72 of 450 individuals (16%), as detected by multigene (panel) testing for inherited cancer susceptibility genes. As expected, the majority of identified variants were in genes known to be associated with CRC, predominantly Lynch syndrome (37 of 72 patients, 51.4%). However, 13 of 72 patients (18.1%) had pathogenic variants in genes not traditionally associated with CRC, including but not limited to BRCA1/BRCA2, which accounted for 8% of the identified variants. Because of the high frequency and wide variety of pathogenic variants identified, the authors suggested consideration of multigene testing for all individuals with early-onset CRC.[344]
Gene-specific considerations and associated CRC risk
The MLH1 and MSH2 genes were originally thought to account for most pathogenic variants of the MMR genes found in Lynch syndrome. However, the prevalence of MSH6 and PMS2 pathogenic variants has been increasing with improved DNA analyses and universal tumor screening of all CRCs.[366] MSH6 and PMS2 variants may be more common in unselected cases of CRC (and endometrial cancer),[366] compared with MLH1 and MSH2 variants which were more commonly identified in individuals from high-risk CRC clinics.[367,368]
MLH1
In early studies, the prevalence of MLH1 pathogenic variants in individuals with Lynch syndrome was reported to be between 41.7% [369] and 50%,[370] making MLH1 the most commonly altered MMR gene in Lynch syndrome families. It was not until a report on the population-based prevalence of Lynch syndrome that the MLH1 pathogenic variant was estimated to be 1 in 1,946, ranking third after PMS2 (1 in 714) and MSH6 (1 in 758), as estimated in a large international study of 5,744 CRC cases.[366]
MLH1 pathogenic variants are associated with the entire spectrum of malignancies associated with Lynch syndrome.[370] The lifetime risk of any Lynch syndrome–associated cancer by age 70 years has been found to range between 59% and 65% in MLH1 pathogenic variant carriers.[285] The highest risk among carriers of pathogenic MLH1 variants is for CRC, which is estimated to be between 41% and 68%,[3,4,365] and the mean age at diagnosis of CRC was 42.8 years (range, 16–81 y) in one study that included 137 affected individuals.[371] In a more recent prospective study using pooled European registry data of 944 MLH1 carriers without cancer, the cumulative CRC incidence was 46% at age 70 years, despite colonoscopic surveillance (albeit at various intervals).[5]
Unlike the APC gene of FAP, in which several phenotypes of differing severity and spectrum of disease occur, genotype-phenotype relationships have been elusive in the MMR genes. In a large series of MLH1 pathogenic variant carriers, women with truncating MLH1 pathogenic variants had significantly later onset of endometrial cancer than did those with nontruncating variants.[372] A nonsignificant trend toward later CRC onset in those with truncating MLH1 pathogenic variants was also identified. As no other correlations were found with specific types of variants in MLH1 or other MMR genes, these associations could be artifactual and warrant further validation.
MSH2
The prevalence of MSH2 pathogenic variants in individuals or families with Lynch syndrome has varied across studies. MSH2 pathogenic variants were reported in 38% to 54% of Lynch syndrome families in studies including large cancer registries and among cohorts of early-onset CRC (younger than age 55 y).[258,373] The reported prevalence of MSH2 pathogenic variants was 32.8% in 2012 in the database of the International Society for Gastrointestinal Hereditary Tumors (InSiGHT), a large professional organization devoted to the collaborative study of familial GI cancer,[369] with families readily ascertained based on the presence of extracolonic cancers in MSH2-associated Lynch syndrome. However, the prevalence of MSH2 pathogenic variants was estimated to be 1 in 2,841 in a population-based cohort of 5,744 CRC cases recruited from the United States, Australia, and Canada;[366] MSH2 was the least prevalent of the MMR gene variants associated with Lynch syndrome.
The risk of any Lynch syndrome–associated cancer by age 70 years has been found to range between 57% to nearly 80% in MSH2 pathogenic variant carriers.[285] The lifetime risk of colon cancer associated with MSH2 pathogenic variants is estimated to be between 48% and 68%.[3,4,365] In a case series of Lynch syndrome patients, those carrying germline MSH2 pathogenic variants (49 individuals, 45% women) had a lifetime (cutoff age, 60 y) risk of extracolonic cancers of 48% compared with 11% for MLH1 carriers (56 individuals, 50% women).[374] In a more recent prospective study using pooled European registry data of 616 MSH2 carriers without cancer, the cumulative CRC incidence was 35% at age 70 years, despite colonoscopic surveillance.[5]
The mean age at diagnosis of CRC in MSH2 carriers has been comparable to MLH1 carriers. One study that included 143 affected individuals with MSH2 pathogenic variants found a mean age at CRC diagnosis of 43.9 years (range, 16–90 y). The same study reported a mean age at CRC diagnosis of 42.8 years (range, 16–81 y) in 137 MLH1 pathogenic variant carriers.[371]
MSH6
Most series have reported a prevalence of germline MSH6 pathogenic variants in approximately 10% of Lynch syndrome families from high-risk clinics and a higher proportion of unselected CRC patients, at approximately 50%.[356,375-380] The reported prevalence of MSH6 pathogenic variants in the InSiGHT database was 18% in 2012.[369] The wide range of prevalence estimates for pathogenic MSH6 variants was a result of small sample sizes, ascertainment bias, and the later age of CRC onset and less striking family histories in MSH6-associated Lynch syndrome families compared with MLH1- and MSH2-associated Lynch syndrome families.[375] This is in line with findings from a population-based study of 42 carriers of deleterious MSH6 germline pathogenic variants, 30 (71%) of whom had a family cancer history that did not meet the Amsterdam II criteria.[6] In a recent, international, population-based study of 5,744 CRC cases, the prevalence of MSH6 pathogenic variants was estimated to be 1 in 758, ranking as the second most prevalent of the MMR genes following PMS2.[366]
The lifetime risk of any Lynch syndrome–associated cancer among MSH6 pathogenic variant carriers is approximately 25% [285] with CRC lifetime risk estimated to be between 12% and 22% [4,6] with MSH6 carriers diagnosed with CRC at a later age than MLH1 and MSH2 carriers. In an earlier study of 146 MSH6 carriers (59 men and 87 women) from 20 families, all of whom had truncating pathogenic variants in MSH6, there was a similar prevalence of CRC by age 70 years among MLH1, MSH2, and MSH6 carriers (P = .0854). However, the mean age at diagnosis for colorectal carcinoma was (a) 55 years for male MSH6 carriers (n = 21; range, 26–84 y) versus 43 years and 44 years in carriers of MLH1 and MSH2 pathogenic variants, respectively; and (b) 57 years for female MSH6 carriers (n = 15; range, 41–81 y) versus 43 years and 44 years in carriers of MLH1 and MSH2 pathogenic variants, respectively.[381]
The largest series of carriers of MSH6 pathogenic variants reported to date includes 113 families from five countries who were ascertained through family cancer clinics and population-based cancer registries.[6] Compared with the incidence for the general population, MSH6 pathogenic variant carriers had an eightfold increased incidence of CRC (HR, 7.6; 95% CI, 5.4–10.8), which was independent of sex and age. By age 70 years, 22% (95% CI, 14%–32%) of male carriers of MSH6 pathogenic variants developed CRC compared with 10% (95% CI, 5%–17%) of female carriers. By age 80 years, the CRC prevalence doubled to 44% (95% CI, 28%–62%) of male carriers of MSH6 pathogenic variants diagnosed with CRC compared with 20% (95% CI, 11%–35%) among female carriers.
In a more recent prospective study using pooled European registry data of 305 MSH6 carriers without cancer, the cumulative CRC incidence was 20% at age 70 years despite colonoscopic surveillance.[5]
PMS2
PMS2 was the last of the genes in the MMR family of genes to be identified. This was because lower penetrance among families made it more difficult to identify [382] using clinical criteria, and also because of limitations of DNA mutational analysis that result from pseudogene interference.
In earlier studies of individuals with CRC and suspected Lynch syndrome, the prevalence of PMS2 pathogenic variants was variable from 2.2% to 5%,[256,383] with an increase to 7.5% as reported in the InSiGHT database in 2012.[369] From a study examining universal tumor testing results from unselected cases of CRC in Switzerland, IHC evaluation of 1,000 consecutive cases found isolated absence of PMS2 expression in 1.5% of all tumors. If this frequency of PMS2-deficient CRCs were representative of all PMS2-associated Lynch syndrome, PMS2 would be the most common gene associated with Lynch syndrome.[384] Results from a large, population-based CRC cohort found that the prevalence of PMS2 pathogenic variants was the highest among all MMR variants, in which 1 person in 714 carried a pathogenic PMS2 gene variant.[366]
The lifetime risk of any cancer has been found to range between 25% and 32% for heterozygous PMS2 pathogenic variant carriers.[285] A meta-analysis of three population-based studies and one clinic-based study estimated that for carriers of PMS2 pathogenic variants, the risk of CRC to age 70 years was 20% among men and 15% among women, and the risk of endometrial cancer was 15%.[385] Similarly, a European consortium of clinic-based registries, taking care to correct for ascertainment bias, found a cumulative lifetime (to age 70 y) CRC risk of only 19% in men and 11% in women with PMS2 pathogenic variants.[386] In addition, patients with PMS2 pathogenic variants presented with CRC 7 to 8 years later than did those with MLH1 and MSH2 pathogenic variants.[383] In a prospective study using pooled European registry data of 77 PMS2 carriers without cancer, the cumulative CRC incidence was 10% at age 70 years despite colonoscopic surveillance.[5] An analysis of nearly 5,000 patients from 284 PMS2 families from the European consortium, supplemented by data from two more registries, was intended to provide more robust PMS2-associated cancer risk estimates.[387] The risk of CRC up to age 80 years was 13% (95% CI, 7.9%–22%) for men and 12% (95% CI, 6.7%–21%) for women, compared with general population risk estimates of 6.6% and 4.7%, respectively. Endometrial cancer risk was found to be 13% (95% CI, 7%–24%). No excess risk of other Lynch syndrome–spectrum tumors was identified in these cohorts. The authors concluded that these data justify consideration of delaying initiation of colonoscopy until age 35 to 40 years, and with longer follow-up intervals (2–3 y), although this was not specifically studied. As with the original reports from the European Prospective Lynch Syndrome Database, it was not possible to assess the extent to which such colonoscopies and polypectomies might have reduced the rate of detected CRCs.
The PLSD is a major ongoing initiative to assess cancer risks in Lynch syndrome. Although it lacks specific details regarding screening practices, it includes outcome data from many European programs, classified by age, gender, and MMR gene.[5,388,389] Recognizing limitations in the larger PLSD, a subset with more detailed surveillance data has been provided.[390] These prospective colonoscopy data from Germany, Holland, and Finland included 2,747 patients of whom 62 had no prior cancer at surveillance initiation. Because of differences in surveillance practices, the colonoscopy interval approximated 1 year in Germany, 2 years in Holland, and 3 years in Finland. The median number of colonoscopies was five and the median per-patient observation time was approximately 8 years. Despite the differences in surveillance intervals, similar adenoma detection rates were found in those patients with a history of cancer (14%) and those without (15.6%). At 10 years of follow-up, rates of first cancer were 8.4% and 14% for metachronous tumors. Factors increasing risk were male sex, prior CRC, presence of MLH1 or MSH2 pathogenic variants, age older than 40 years, and adenoma at index colonoscopy. Notably, no significant difference in CRC detection or in stage at detection was noted between screening intervals of 1, 2, or 3 years.
It is important to note that a more severe phenotype is seen among carriers of biallelic PMS2 pathogenic variants. For more information, see the IHC in constitutional mismatch repair deficiency (CMMRD) syndrome section.
The lifetime risk of CRC and endometrial cancer in carriers of these pathogenic variants is summarized in Table 11.
| Gene | Lifetime Risk of Colorectal Cancer (%) | Lifetime Risk of Endometrial Cancer (%) | References |
|---|---|---|---|
| MLH1 | 41–50 | 34–54 | [3-5] |
| MSH2 | 35–56 | 21–51 | [3-5] |
| MSH6 | 10–22 | 16–49 | [4-6] |
| PMS2 | 10 | 24 | [5] |
EPCAM
A subset of individuals with Lynch syndrome (approximately 1%) have a pathogenic variant in EPCAM, which leads to hypermethylation and inactivation of the MSH2 promoter.[391] In a European study of 194 EPCAM deletion carriers, the cumulative risk of CRC up to age 70 years was 75% with the average age at onset of 43 years. This is comparable to the risk in MSH2 carriers (up to 68% by age 70 y). However, the risk of endometrial cancer among women with an EPCAM deletion was only 12% in this study, compared with a risk of up to 71% in MSH2 carriers.[392] The associated phenotype is dependent on the location of the deletion variant in the 3’ end of the EPCAM gene; if the deletion is large and includes parts of the promoter of MSH2, the phenotype will be similar to other MSH2-associated Lynch syndrome families.[392] When the deletion involves the termination signal of EPCAM but spares all of the MSH2 gene and promoter, the phenotype is mainly confined to CRC.[393]
One study of two families with the same EPCAM deletion limited to the 3’ end of the gene and not extending into the promoter of MSH2 found few extracolonic cancers and no endometrial cancers.[393] However, a subsequent study demonstrated that women with MSH2 protein expression loss caused by EPCAM variants are also at risk of endometrial cancer.[392]
Intragenic variability in CRC penetrance among carriers of MMR pathogenic variants
While it is known that MMR pathogenic variant carriers have high lifetime risks of CRC, estimates of these risks vary considerably between studies. Additional factors, including ascertainment bias, heritable modifiers of risk, and nonheritable risk factors may contribute substantially to variation in lifetime risk estimates.
A study from the Colon Cancer Family Registry reported that, depending on the gene that has the pathogenic variant and an individual's sex, 16% to 23% of MLH1 and MSH2 carriers had lifetime CRC risks of less than 10% (i.e., their risks approached average risk for the general population).[394] Conversely, 10% to 17% of carriers had lifetime risks of more than 90% (i.e., these carriers were almost certain to develop CRC).
An international registry–based study used retrospective data to examine variation in CRC risk in 5,255 families with Lynch syndrome.[395] The study used a segregation analysis conditioned on ascertainment to estimate mean CRC penetrance and modeled unmeasured polygenic factors to estimate variations in penetrance. Nearly 80,000 individuals were included in the study. The number of patients with EPCAM pathogenic variants was insufficient to include in the analysis. Therefore, only families with MLH1, MSH2, MSH6 and PMS2 pathogenic variants were included. Patients were clustered by continent (North America, Europe, and Australasia). There was strong evidence that unknown familial risk factors modified CRC risk for Lynch syndrome carriers across all continents. This resulted in wide variation in CRC risk for each MMR gene. This variation was most pronounced for MLH1 and MSH2, in which 7% to 56% of carriers had a CRC penetrance of less than 20%, 9% to 44% of carriers had a CRC penetrance of more than 80%, and only 10% to 19% of carriers had a CRC penetrance of 40% to 60%. Wide variation in CRC risk was observed even when the analysis was restricted to the 250 families carrying the most common variant in the dataset (the MSH2 pathogenic c.942+3A>T variant). In this MSH2 cohort, 9% to 15% of carriers had a CRC penetrance of less than 20%, and 33% to 45% of carriers had a CRC penetrance of more than 80%. Limitations of the study included the following: (1) 94% of patients were ascertained from high-risk clinics, (2) there were limited data on screening and polypectomy, and (3) multiple assumptions and models were used. These results suggest that published mean cancer risks may not be representative for most Lynch syndrome carriers. Therefore, the authors suggested that current screening guidelines might not be applicable to large proportions of Lynch syndrome patients. Until additional studies have identified and characterized genetic and environmental risk modifiers, contemporary guidelines are recommended for most Lynch syndrome carriers.
CMMRD
As described above, patients may carry MMR gene variants in both parental alleles, in a condition known as CMMRD. For more information, see the IHC in constitutional mismatch repair deficiency (CMMRD) syndrome section.
The occurrence of such biallelic variants is associated with a characteristic but not diagnostic clinical phenotype. Clinical features include hematologic malignancies and brain tumors in children. When GI tumors occur, the age of onset is strikingly low, sometimes before age 20 years. Café au lait spots and features otherwise suggesting neurofibromatosis are characteristic. Occasionally, patients present with multiple adenomas.
Ethnic variation and founder pathogenic variants in Lynch syndrome
The frequency of MMR variants does not differ markedly from population to population, with similar frequencies identified in a host of different countries. As with hereditary breast and ovarian cancer (HBOC), there are certain variants that occur at higher frequencies within a particular ethnic group. Notable in HBOC are the commonly recurring Ashkenazi Jewish variants, so common that direct-to-consumer testing is offered for these common variants. (Refer to the Prevalence of BRCA1/2 Pathogenic Variants section in BRCA1 and BRCA2: Cancer Risks and Management and the Direct-to-Consumer [DTC] Genetic Tests section in Cancer Genetics Risk Assessment and Counseling for more information.) The ancientness of apparent founder variants is generally established by haplotype analysis. In some instances, what may appear to be a founder variant is simply a frequently recurring de novo variant.[396]
Among the first population findings regarding the MMR genes of Lynch syndrome was the recognition of two very common MLH1 variants in Finland, accounting for most cases of Lynch syndrome in this country.[397,398] Since that time, founder variants have been identified in most populations in which relatively unselected series of patients with CRC have undergone variant testing. Many of the reports originate in Europe. As in Finland, these may be straightforward to identify in the setting of fairly homogeneous ethnicity with low immigration. Founder variants in Europe have been found in the United Kingdom, Sweden, Switzerland, Italy,[399] Portugal, France, Spain, and Hungary, and are likely present in all ethnic groups. Fewer such reports have come from Asia,[400] Latin America, the Middle East, and Africa.
In the United States, a deletion in exons 1–6 of the MSH2 gene has been estimated to account for as much as 20% of variants in that gene. This so-called American Founder Mutation has been determined by haplotype analysis to date back about 500 years.[401]
A South American study combining data from Uruguay, Colombia, Brazil, Argentina, and Chile also selected cases of interest according to Amsterdam and Bethesda features, yielding a 60% frequency of MLH1 and 40% frequency of MSH2. MSH6 and PMS2 were not evaluated. Selection bias likely influenced the frequency of variants and perhaps the relative contributions by MLH1 and MSH2. A possible founder variant in Colombia was noted.[402]
Although testing for commonly recurring founder variants in a given ethnic/geographic area has been considered to be a cost-effective first step when a stepwise strategy is employed, it is likely not necessary when the increasingly common approach of broad panel testing is undertaken as a basic strategy.
Some ethnic groups have increased rates of consanguinity and a subsequent risk of CMMRD. For more information, see the IHC in constitutional mismatch repair deficiency (CMMRD) syndrome section.
Ethnic variation in the United States
In this section, the data exploring the distribution of MMR gene variants amongst differing ethnic groups in the United States are presented. The interpretation of these studies is challenging given the presence of selection and ascertainment bias. In addition, even population-based studies are limited by small sample sizes for many ethnic groups and self-reporting of ethnicity/race.
There are few data suggesting the presence of much variation in Lynch syndrome frequency according to geography or ethnicity. Within a small and/or homogeneous ethnic group the presence of founder variants may seem to increase the prevalence of variants in that particular gene. Slight differences in the proportion of MLH1 and MSH2 variants exist from one population to another. MSH6 and PMS2 have been insufficiently studied at the population level as to enable inferences about their relative frequencies.
The most representative population-based studies in the United States, such as that in Columbus, Ohio, have been overrepresented by White individuals, in accordance with their greater overall numbers. Consequently, data on racial and ethnic minorities such as Hispanic and African American individuals suffer from smaller and less rigorously representative samples.
A study conducted in Puerto Rico considered variants in 89 Caribbean Hispanic patients with Lynch syndrome suspected on the grounds of Amsterdam criteria or Bethesda guidelines.[403] Patients underwent either immediate germline testing or step-wise evaluation beginning with tumor MSI/IHC. Frequencies of variants by gene were 67% for MSH2, 25% for MLH1, and 8% for MSH6. No definite founder variants were evident. Clearly, the selection of participants according to clinical family history criteria would have led to an underreporting of the less penetrant MSH6 and PMS2 genes.
Clinic-based series from California, Texas, and Puerto Rico yielded an overall variant prevalence similar to those described, with somewhat more MLH1 than MSH2, but also including MSH6 and PMS2. Presence of potential founder variants traceable back to Spain and Europe were noted.[404]
The closest population-based information on Lynch syndrome in Hispanic individuals is a Southern California study based on the California Tumor Registry, in which 265 patients were identified.[405] Of those with MSI-H tumors, 13 (62%) had MMR variants. Frequencies of MMR variants were 46% for MLH1 (6 of 13), 31% for MSH2 (4 of 13), 15% for MSH6 (2 of 13), and 8% for PMS2 (1 of 13).
The problem of small numbers is highlighted by the findings from the more truly population-based studies that have been done in the United States. In a study from Columbus, Ohio, only 8% of the consecutive series patients were African American and the proportion of Hispanic individuals as a subset of White individuals was not stated.[344] In another study involving panel testing of nearly all CRC patients treated at Dana-Farber Cancer Institute, less than 5% were Black and less than 3% were Hispanic, underscoring the challenge of extracting meaningful data from small subsets.[347]
Lynch syndrome in African Americans
The issues in evaluating prevalence of Lynch syndrome and cancer risks associated with MMR variants in African American individuals are similar to those in Hispanic individuals: a heterogeneous population that has been understudied. A study of clinic-based data from 13 referral centers in the United States identified 51 families with Lynch syndrome with frequencies of MMR gene variants as follows: 61% MLH1, 21% MSH2, 6% MSH6, and 12% PMS2. Age of cancer onset distribution curves were very similar to those seen in White populations.[406] As with most of the studies in Hispanic individuals, cases were not identified according to any consistent, programmatic evaluation such as universal tumor testing.
Risk of metachronous CRC
A hallmark feature of Lynch syndrome is that carriers of pathogenic MMR gene variants have an increased risk of development of synchronous and metachronous colorectal neoplasms.[388,407,408] In one study of 382 individuals with Lynch syndrome from the Colon Cancer Family Registry, the incidence of metachronous CRCs was 16% at 10 years, 41% at 20 years, and 63% at 30 years after segmental colectomy.[407] The risk of metachronous CRC decreased in a stepwise fashion by 31% for every 10 cm of the colon that was removed, with none of the 50 individuals who had extensive colectomies diagnosed with metachronous CRC. Another prospective study of 1,273 patients with Lynch syndrome who had prior cancer reported a cumulative incidence of subsequent CRC of 46% for MLH1 carriers, 48% for MSH2 carriers, and 23% for MSH6 carriers. This represents only a slightly greater risk of new cancers than pathogenic variant carriers with no previous cancer diagnosis. Excellent survival was again seen and was regarded as a combination of favorable tumor pathology and the effect of surveillance.[388]
Risk of extracolonic malignancies associated with Lynch syndrome
Patients with Lynch syndrome are at an increased risk of other cancers, especially those of the endometrium. The cumulative risk of extracolonic cancer has been estimated to be 20% by age 70 years in 1,018 women in 86 families, compared with 3% in the general population.[409] There is some evidence that the rate of individual cancers varies from kindred to kindred.[410-412]
Endometrial cancer
The most common extracolonic malignancy in Lynch syndrome is endometrial adenocarcinoma, which affects at least one female member in about 50% of Lynch syndrome families. In addition, 50% of women with an MMR gene pathogenic variant will present with endometrial cancer as her first malignancy.[413]
The lifetime risk of endometrial cancer has been estimated to be from 44% in carriers of MLH1 pathogenic variants to 71% in carriers of MSH2 pathogenic variants, although some earlier studies may have overestimated risk due to ascertainment bias.[6,260,365,373,414] Lifetime risk of endometrial cancer in carriers of MSH6 pathogenic variants in 113 families was estimated to be 26% at age 70 years and 44% at age 80 years;[6] overall, female carriers of MSH6 pathogenic variants had an endometrial cancer risk that was 25 times higher than women in the general population (HR, 25.5; 95% CI, 16.8–38.7; P < .001).[6] In another study, the cumulative lifetime risk of uterine cancer was higher in MSH6 carriers (71%) than in carriers of MLH1 (27%) and MSH2 (40%) pathogenic variants (P = .02), with an older mean age at diagnosis of 54 years in carriers of MSH6 pathogenic variants (n = 29; range, 43–65 y) versus 48 years in carriers of MLH1 and 49 years in carriers of MSH2 pathogenic variants.[381] In carriers of PMS2 pathogenic variants, the endometrial cancer risk at age 70 years has been reported to be 15%.[385] Prospective data collected in the Colon Cancer Family Registry program yielded 5-year endometrial cancer risks of about 3% and 10-year endometrial cancer risks of about 10% among women with MMR gene pathogenic variants.[415] A prospective study using pooled European registry data of 1,942 MMR carriers without prior cancer reported a cumulative incidence of endometrial cancer of 34% in MLH1 carriers, 51% in MSH2 carriers, 49% in MSH6 carriers, and 24% in PMS2 carriers.[5] Women with loss of MSH2 protein expression caused by an EPCAM pathogenic variant are also at risk of endometrial cancer depending upon the location of the variant in EPCAM. One study found a 12% (95% CI, 0%–27%) cumulative risk of endometrial cancer in EPCAM deletion carriers.[392]
A study of 127 women with Lynch syndrome who had endometrial cancer as their index cancer were found to be at significantly increased risk of other cancers. The following elevated risks were reported: CRC, 48% (95% CI, 27.2%–58.3%); kidney, renal pelvis, and ureter cancer, 28% (95% CI, 11.9%–48.6%); urinary bladder cancer, 24.3% (95% CI, 8.56%–42.9%; and breast cancer, 2.51% (95% CI, 1.17%–4.14%).[416]
In a study of 113 families that carried MSH6 pathogenic variants from the Colon Cancer Family Registry, female MSH6 carriers had a 26-fold increased incidence of endometrial cancer (HR, 25.5; 95% CI, 16.8–38.7) compared with the general population. A sixfold increased incidence of other cancers associated with Lynch syndrome (HR, 6.0; 95% CI, 3.4–10.7) was observed compared with the general population, but not among male MSH6 carriers.[6]
Lynch syndrome–associated endometrial cancer is not limited to the endometrioid subtype, and the spectrum of uterine tumors in Lynch syndrome may include clear cell carcinoma, uterine papillary serous carcinoma, and malignant mixed Müllerian tumors.[417] Also, endometrial cancer most commonly arises from the lower uterine segment. (Refer to the Endometrial cancer screening in Lynch syndrome section of this summary for information about screening methods.)
Cancer risk in Lynch syndrome beyond CRC and endometrial cancer
Multiple studies demonstrate an increased risk of additional malignancies associated with Lynch syndrome, including cancers of the stomach, pancreas, ovary, small intestine, and brain, transitional cell carcinoma of the bladder, ureters, and renal pelvis, and sebaceous adenomas of the skin.[409,410,418-421] In addition, some studies have suggested an association with breast, prostate, and adrenal cortex cancers.[415,419,422-424] The strength of the association for many of these malignancies is limited by the majority of studies having a small sample size (and consequently, wide CIs associated with relative risk [RR]), the retrospective nature of the analyses, and referral or ascertainment bias.
The largest prospective study to date is of 446 unaffected carriers of pathogenic variants from the Colon Cancer Family Registry.[415] The Colon Cancer Family Registry is an international cohort with both population-based and clinic-based recruitment from six centers in North America and Australia. Control subjects were noncarriers from families with a known MMR pathogenic variant. Three subcohorts were used to analyze the risk of CRC (365 carriers, 903 noncarriers), endometrial cancer (215 carriers, 523 noncarriers), and other cancers (446 carriers, 1,029 noncarriers). Participants who were followed for up to 10 years demonstrated an increased SIR for CRC (SIR, 20.48; 95% CI, 11.71–33.27; P < .01), endometrial cancer (SIR, 30.62; 95% CI, 11.24–66.64; P < .001), ovarian cancer (SIR, 18.81; 95% CI, 3.88–54.95; P < .001), gastric cancer (SIR, 9.78; 95% CI, 1.18–35.30; P = .009), renal cancer (SIR, 11.22; 95% CI, 2.31–32.79; P < .001), bladder cancer (SIR, 9.51; 95% CI, 1.15–34.37; P = .009), pancreatic cancer (SIR, 10.68; 95% CI, 2.68–47.70; P = .001), and female breast cancer (SIR, 3.95; 95% CI, 1.59–8.13; P = .001).[415]
A well-described variant of Lynch syndrome whose phenotype includes multiple cutaneous neoplasms (including sebaceous adenomas, sebaceous carcinomas, and keratoacanthomas) and CRC is Muir-Torre syndrome.[425,426] Pathogenic variants in the MLH1, MSH2, and MSH6 genes have been found in Muir-Torre families with an increased prevalence described among MSH2 carriers.[427-434] A study of 1,914 unrelated MLH1 and MSH2 probands found MSH2 to be more common in individuals with the Muir-Torre syndrome phenotype. Of 15 individuals with sebaceous skin tumors, 13 (87%) had MSH2 pathogenic variants compared with two individuals who had MLH1 pathogenic variants (P = .05).[435] Evidence of defective DNA MMR activity using IHC or MSI testing was reported in 69 of 163 randomly collected sebaceous neoplasms (42%), suggesting that this is a common mechanism for the development of these lesions, and that testing for defective MMR in sebaceous neoplasms would be an ineffective means to screen for Lynch syndrome or Muir-Torre syndrome.[436] (Refer to the Sebaceous Carcinoma section in the PDQ summary on Genetics of Skin Cancer for more information about cutaneous neoplasms in Muir-Torre syndrome.)
| Cancer Siteb | General Population Risk (%)c | Risk in Individuals With Lynch Syndrome (%)d | References |
|---|---|---|---|
| CNS = central nervous system. | |||
| aAdapted from Syngal et al.[437] | |||
| bEvolving data suggest a potential association between Lynch syndrome and breast and prostate cancers. (Refer to the Additional cancers potentially associated with Lynch syndrome section of this summary for more information about these cancers.) | |||
| cHowlader et al.[438] | |||
| dRange of cancer risk estimates vary based on study sample size, subject ascertainment, and statistical methods. | |||
| Stomach | <1 | 0.2–13 | [4,6,371,439-444] |
| Ovary | 1.3 | 3.4–22 | [4,6,365,371,381,385,440-442,444,445] |
| Hepatobiliary tract | <1 | 0.02–4 | [4,442,444-446] |
| Urinary tract | <1 | 0.2–25.5 | [4,371,385,440-442,444,445,447] |
| Small bowel | <1 | 0.4–12 | [4,343,371,439-442] |
| Brain/CNS | <1 | 1.2–3.7 | [371,440,442,444] |
| Sebaceous neoplasms | <1 | 9.0 | [429,448,449] |
| Pancreas | 1.6 | 0.4–3.7 | [440,450,451] |
Additional cancers potentially associated with Lynch syndrome
Additional tumors are being considered as part of the spectrum of Lynch syndrome, but this is controversial. Breast and prostate cancers have been raised as possible Lynch syndrome–associated tumors such that MMR genes are now included on multigene (panel) tests for these cancers.
Breast cancer
The issue of breast cancer risk in Lynch syndrome has been controversial.
Retrospective studies have been inconsistent, but several have demonstrated microsatellite instability in a proportion of breast cancers from individuals with Lynch syndrome;[452-455] one of these studies evaluated breast cancer risk in individuals with Lynch syndrome and found that it is not elevated.[455] However, the largest prospective study to date of 446 unaffected carriers of pathogenic variants from the Colon Cancer Family Registry [415] who were followed for up to 10 years reported an elevated SIR of 3.95 for breast cancer (95% CI, 1.59–8.13; P = .001).[415] The same group subsequently analyzed data on 764 carriers of MMR gene pathogenic variants with a prior diagnosis of colorectal cancer. Results showed that the 10-year risk of breast cancer following colorectal cancer was 2% (95% CI, 1%–4%) and that the SIR was 1.76 (95% CI, 1.07–2.59).[456] A series from the United Kingdom composed of clinically referred Lynch syndrome kindreds, with efforts to correct for ascertainment, showed a twofold increased risk of breast cancer in 157 MLH1 carriers but not in carriers of other MMR variants.[457] Results from a meta-analysis of breast cancer risk in Lynch syndrome among 15 studies with molecular tumor testing results revealed that 62 of 122 breast cancers (51%; 95% CI, 42%–60%) in MMR pathogenic variant carriers were MMR-deficient. In addition, breast cancer risk estimates among a total of 21 studies showed an increased risk of twofold to 18-fold in eight studies that compared MMR variant carriers with noncarriers, while 13 studies did not observe statistical evidence for an association of breast cancer risk with Lynch syndrome.[458]
A number of subsequent studies have suggested the presence of higher breast cancer risks than previously published,[348,349,459,460] although this has not been consistently observed.[341] Through a study of 325 Canadian families with Lynch syndrome, primarily encompassing MLH1 and MSH2 carriers, the lifetime cumulative risk for breast cancer among MSH2 carriers was reported to be 22%.[459] Similarly, breast cancer risks were elevated in a study of 423 women with Lynch syndrome, with substantially higher risks among those with MSH6 and PMS2 pathogenic variants, compared with MLH1 and MSH2 pathogenic variants.[349] In fact, breast cancer risk to age 60 years was 37.7% for PMS2, 31.1% for MSH6, 16.1% for MSH2, and 15.5% for MLH1. These findings are consistent with another study of 528 patients with Lynch syndrome–associated pathogenic variants (including MLH1, MSH2, MSH6, PMS2, and EPCAM) in which PMS2 and MSH6 variants were much more frequent among patients with only breast cancer, compared with those with only colorectal cancer (P = 2.3 x 10-5).[348] Additional data to support an association of MSH6 with breast cancer were provided through a study of over 10,000 cancer patients across the United States who had genetic testing.[460] Findings indicated that MSH6 was associated with breast cancer with an odds ratio (OR) of 2.59 (95% CI, 1.35–5.44). Taken together, these studies highlight how the risk profile among patients with Lynch syndrome is continuing to evolve as more individuals are tested through multigene panel testing, with representation of larger numbers of individuals with PMS2 and MSH6 pathogenic variants compared with prior studies. In the absence of definitive risk estimates, individuals with Lynch syndrome are screened for breast cancer on the basis of family history.[461]
Prostate cancer
Prostate cancer was found to be associated with Lynch syndrome in a study of 198 families from two U.S. Lynch syndrome registries in which prostate cancer had not originally been part of the family selection criteria. Prostate cancer risk in relatives of carriers of MMR gene pathogenic variants was 6.3% at age 60 years and 30% at age 80 years, versus a population risk of 2.6% at age 60 years and 18% at age 80 years, with an overall HR of 1.99 (95% CI, 1.31–3.03).[422] A 2014 meta-analysis supports this association, finding an estimated RR of 3.67 (95% CI, 2.32–6.67) for prostate cancer in men with a known MMR pathogenic variant.[462] This risk is possibly increased in those with MSH2 pathogenic variants.[424,462] Notwithstanding prevalent controversy surrounding routine prostate-specific antigen (PSA) screening, the authors suggested that screening by means of PSA and digital rectal exam beginning at age 40 years in male MMR gene carriers would be “reasonable to consider.”[422] A study of 692 men with metastatic prostate cancer unselected for family history of cancer or age at diagnosis identified germline MMR pathogenic variants in four men (0.5%).[463] Currently, molecular and epidemiological evidence supports prostate cancer as one of the Lynch syndrome cancers. As with breast cancer,[462] additional studies are needed to define absolute risks and age distribution before surveillance guidelines for prostate cancer can be developed for carriers of MMR pathogenic variants. For more information about prostate cancer and Lynch syndrome, see the DNA mismatch repair genes (Lynch syndrome) section in Genetics of Prostate Cancer.
Adrenocortical cancer
In a series of 114 ACC cases, of which 94 patients had a detailed family history assessment and Li-Fraumeni syndrome was excluded, three patients had family histories that were suggestive of Lynch syndrome. The prevalence of MMR gene pathogenic variants in 94 families was 3.2%, similar to the proportion of Lynch syndrome among unselected colorectal and endometrial cancer patients. In a retrospective review of 135 MMR gene pathogenic variant–positive Lynch syndrome families from the same program, two probands were found to have had a history of ACC. Of the four ACCs in which MSI testing could be performed, all were MSS. These data suggest that if Lynch syndrome is otherwise suspected in an ACC index case, an initial evaluation of the ACC using MSI or IHC testing may be misleading.[423]
Other cancers
Several additional cancers have been found to be associated with Lynch syndrome in some studies, but further investigation is warranted. Table 12 compares the risk of these cancers in the general population with that of individuals with Lynch syndrome.
Management of Lynch syndrome
Screening and surveillance in Lynch syndrome
Colon cancer screening and surveillance in Lynch syndrome
Several aspects of the biologic behavior of CRC and its precursor lesion, the adenomatous polyp, in individuals with Lynch syndrome support a different approach to CRC screening in this population as compared with those recommendations for average-risk people in the general population. At present, the recommendations for cancer screening and surveillance in Lynch syndrome consider the differences in cancer risks as compared with those in the general population due to the causative germline deficiency in the MMR system. The following biological differences form the basis of the currently implemented screening strategies in Lynch syndrome:
-
CRC and adenomas present at a younger age.
CRCs in Lynch syndrome occur earlier in life than do sporadic cancers; however, the age of onset varies based on which of the MMR genes is altered. (Refer to the Prevalence, clinical manifestations, and cancer risks associated with Lynch syndrome section of this summary for more information about gene-specific age of onset of CRC.)
Carriers of Lynch syndrome pathogenic variants have an increased risk of developing colon adenomas and the onset of adenomas appears to occur at a younger age than in pathogenic variant–negative individuals from the same families.[464] The risk of a carrier of MMR pathogenic variants developing adenomas has been reported to be 3.6 times higher than the risk in noncarriers.[464] By age 60 years, 70% of the carriers developed adenomas, compared with 20% of noncarriers. Most of the adenomas in carriers had absence of MMR protein expression and were more likely to have dysplastic features, compared with adenomas from control subjects.[464]
In one study, the mean age at diagnosis of adenoma in carriers was 43.3 years (range, 23–63.2 y), and the mean age at diagnosis of carcinoma was 45.8 years (range, 25.2–57.6 y).[464]
-
There is a right-sided predominance of colon cancer.
A larger proportion of Lynch syndrome CRCs (60%–70%) occur in the right colon, suggesting that sigmoidoscopy alone is not an appropriate screening strategy and that a colonoscopy provides a more complete structural examination of the colon. Evidence-based reviews of surveillance colonoscopy in Lynch syndrome have been reported.[125,465,466] The incidence of CRC throughout life is substantially higher in patients with Lynch syndrome, suggesting that the most-sensitive test available should be used. (Refer to Table 13 for available colon surveillance recommendations.)
-
The adenoma-carcinoma sequence is accelerated.
The progression from normal mucosa to adenoma to cancer is accelerated,[467,468] suggesting that screening should be performed at shorter intervals (every 1–2 years) and with colonoscopy.[468-471] It has been demonstrated that carriers of MMR gene pathogenic variants develop detectable adenomas at an earlier age than do noncarriers.[464,464] It is not known whether this reflects a greater prevalence of adenomas or the presence of larger adenomas with better detection in Lynch syndrome.
Evidence for the use of colonoscopy for CRC screening and surveillance in Lynch syndrome
The risk of CRC in Lynch syndrome has been studied and updated in a Finnish screening trial, which spans from the early 1980s to present.[468,472] Over the course of this trial, the design of the longitudinal study has evolved. In the earliest period, information about each individual's variant status was unknown and study participants were eligible based on fulfillment of clinical criteria; the study consisted of some people with a previous cancer or adenoma diagnosis and others without such history who were undergoing asymptomatic screening while the comparison group was composed of individuals from those same families who refused screening. Many of these people (68%) had screening with x-ray contrast/barium enema. Colonoscopy was the approach used for carriers of MMR pathogenic variants when this information was obtainable and the interval between exams was shortened from 5 years to 3 years to 2 years, based on results from the study over time.
A 15-year controlled screening trial conducted in this series demonstrated a reduction in the incidence of CRC, CRC-specific mortality, and overall mortality with colonoscopy in individuals from Lynch syndrome families.[468] Colonic screening was provided at 3-year intervals in 133 individuals from Lynch syndrome families and 119 controls from these families had no screening. Among those screened, 8 individuals (6%) developed CRC compared with 19 control subjects (16%), for a risk reduction of 62% with screening. Furthermore, all CRCs in the screened group were local, causing no deaths, while there were 9 deaths caused by CRC in the control group. There was also a benefit in overall mortality in the screened group with 10 deaths in the screened group and 26 deaths in the control group (P = .003).
The series subsequently limited its attention to subjects without prior diagnosis of adenoma or cancer. The eligible 420 carriers of pathogenic variants had a mean age of 36 years and underwent an average of 2.1 colonoscopies, with a median follow-up of 6.7 years. Adenomas were detected in 28% of subjects. Cumulative risk of one or more adenomas by age 60 years was 68.5% in men and 48.3% in women. Notably, risk of detecting cancer in those free of cancer at baseline exam, and thus regarded as interval cancers, by age 60 years was 34.6% in men and 22.1% in women. The combined cumulative risk of adenoma or cancer by age 60 years was 81.8% in men and 62.9% in women. For both adenomas and carcinomas, about one-half were located proximal to the splenic flexure. While the rates for CRC despite colonoscopy surveillance appear high, the recommended short intervals were not regularly adhered to in this nonrandomized series. These authors recommended surveillance at 2-year intervals. This is in line with most consensus guidelines (refer to Table 13), in which the appropriate colonoscopy screening interval remains every 1 to 2 years. Analysis of colonoscopic surveillance data in 242 carriers of pathogenic variants 10 years after testing shows 95% compliance in surveillance procedures for CRC and endometrial cancer. Although not all CRCs were prevented, mortality was comparable with variant-negative relatives. However, this may be attributable to the modest sample size of the study.[472]
Individuals with Lynch syndrome are at an increased risk of developing synchronous CRC. Of 5,304 CRC cases in the Danish HNPCC Register, including 774 with Lynch syndrome, the relative risks of synchronous CRC (>1 CRC) diagnosed within 1 year of primary CRC for Lynch syndrome, familial CRC (cases meeting Amsterdam I or II criteria) and metachronous CRC (1 CRC at age <50 y or >2 CRCs at age >50 y) were 5.6, 3.2, and 1.9, respectively, compared with sporadic CRC. Thus, the increased risk of synchronous CRC in patients with a strong family history of CRC, and especially Lynch syndrome, should be considered in preoperative colonoscopic examinations.[473]
Given that colonoscopy is the accepted measure for colon cancer surveillance, preliminary data suggest that the use of chromoendoscopy, such as with indigo carmine, may increase the detection of diminutive, histologically advanced adenomas.[474,475]
When an adenoma is detected, the question of whether to test the adenoma for MSI/IHC is raised. One study of patients with prior CRC and known MMR pathogenic variants found eight of 12 adenomas to have both MSI and IHC protein loss.[476] However, the study authors emphasized that normal MSI/IHC testing in an adenoma does not exclude Lynch syndrome. Abnormal MSI/IHC are uncommon in the smallest adenomas, and more prevalent in adenomas larger than 8 mm, which also suggests that the MMR defect is acquired in the growing adenoma.[477]
Special considerations: The impact of gene-specific variability in cancer risk on CRC screening recommendations in Lynch syndrome
Because of the variability of gene-specific CRC risks, experts in the field have proposed gene-specific screening and surveillance recommendations. For example, a European consortium [386] made a clinical recommendation for delaying the onset of colorectal and endometrial cancer screening to age 30 years, in line with their recommendation for later initiation of screening for carriers of MSH6 pathogenic variants. Additionally, a 2015 review by an ad hoc American virtual workgroup involved in the care of Lynch syndrome patients and families concluded that despite multiple studies indicating reduced penetrance in monoallelic PMS2 carriers, they could not recommend any changes to Lynch syndrome cancer surveillance guidelines for this group.[382]
Available recommendations for colon surveillance in individuals with Lynch syndrome are summarized in Table 13. Most organizations tailor surveillance recommendations for each specific gene.[123,478-480] All of the screening recommendations assume findings are normal. A more aggressive screening schedule might be considered on an individualized basis.
| Organization | MLH1 | MSH2 | MSH6 | PMS2 | EPCAM |
|---|---|---|---|---|---|
| CRC = colorectal cancer; EHTG = European Hereditary Tumor Group; ESCP = European Society of Coloproctology; ESMO = European Society for Medical Oncology; MMR = mismatch repair; NCCN = National Comprehensive Cancer Network. | |||||
| aThis table summarizes available guidelines from 2014 and later. Other organizations, including the American Cancer Society, have published guidelines before 2014.[481] | |||||
| bU.S. Multi-Society Task Force on Colorectal Cancer includes the following organizations: American Academy of Family Practice, American College of Gastroenterology, American College of Physicians-American Society of Internal Medicine, American College of Radiology, American Gastroenterological Association, American Society of Colorectal Surgeons, and American Society for Gastrointestinal Endoscopy. | |||||
| cConsider later age for MSH6 carriers.[480] | |||||
| dConsider repeating colonoscopy every 5 years for PMS2 carriers.[479] | |||||
| eConsider starting at age 30 for MSH6 carriers and 35 for PMS2 carriers. Consider annual colonoscopy for MMR carriers.[316] | |||||
| NCCN (2024) [123] | High-quality colonoscopy at age 20–25 y or 2–5 y prior to earliest CRC in the family if it was diagnosed before age 25 y; repeat colonoscopy every 1–2 y | High-quality colonoscopy at age 20–25 y or 2–5 y prior to earliest CRC in the family if it was diagnosed before age 25 y; repeat colonoscopy every 1–2 y | High-quality colonoscopy at age 30–35 y or 2–5 y prior to earliest CRC in the family if it was diagnosed before age 30 y; repeat colonoscopy every 1–3 y | High-quality colonoscopy at age 30–35 y or 2–5 y prior to earliest CRC in the family if it was diagnosed before age 30 y; repeat colonoscopy every 1–3 y | NCCN recommends that EPCAM carriers be managed the same as MSH2 carriers. High-quality colonoscopy at age 20–25 y or 2–5 y prior to the earliest CRC in the family if it was diagnosed before age 25 y; repeat colonoscopy every 1–2 y |
| ESMO (2020) [480] | Colonoscopy at age 25 y or 5 y prior to earliest CRC if diagnosed before age 25 y; repeat every 1–2 y | Colonoscopy at age 25 y or 5 y prior to earliest CRC if diagnosed before age 25 y; repeat every 1–2 y | Colonoscopy at age 35 y or 5 y prior to earliest CRC if diagnosed before age 25 yc; repeat every 1–2 y | Colonoscopy at age 35 y or 5 y prior to earliest CRC if diagnosed before age 25 y; repeat every 1–2 y | Not addressed |
| British Society of Gastroenterology (BSG)/ Association of Coloproctology of Great Britain and Ireland (ACPGBI)/ United Kingdom Cancer Genetics Group (UKCGG) (2020) [478] | Colonoscopy at age 25 y; repeat every 2 y until age 75 y | Colonoscopy at age 25 y; repeat every 2 y until age 75 y | Colonoscopy at age 35 y; repeat every 2 y until age 75 y | Colonoscopy at age 35 y; repeat every 2 y until age 75 y | EPCAM carriers should be managed as those with MSH2 pathogenic variants |
| European guidelines from the EHTG and ESCP; updated Mallorca group guidelines (2021) [479] | Colonoscopy at age 25 y; repeat every 2–3 y | Colonoscopy at age 25 y; repeat every 2–3 y | Colonoscopy at age 35 y; repeat every 2–3 y | Colonoscopy at age 35 y; repeat every 2–3 yd | Not addressed |
| U.S. Multi-Society Task Force on Colorectal Cancer (2014)b [316] | Colonoscopy beginning at age 20–25 y for 2–5 y prior to earliest CRC if before age 25 y; repeat every 1–2 ye | ||||
Extracolonic cancer screening in Lynch syndrome
Endometrial cancer screening in Lynch syndrome
Note: A separate PDQ summary on Endometrial Cancer Screening in the general population is also available.
Cancer of the endometrium is the most common extracolonic cancer observed in Lynch syndrome families, affecting at least one female in about 50% of Lynch syndrome families. (Refer to the Endometrial cancer section of this summary for more information about gene-specific risks of endometrial cancer in carriers of MMR pathogenic variants.)
In the general population, the diagnosis of endometrial cancer is generally made when women present with symptoms like abnormal or postmenopausal bleeding. Endometrial sampling is performed to provide a histological specimen for diagnosis. Eighty percent of women with endometrial cancer present with stage I disease, and there are no data to suggest that the clinical presentation in women with Lynch syndrome differs from that in the general population.
Given their substantial increased risk of endometrial cancer, endometrial cancer screening has been suggested for women with Lynch syndrome who have not had risk-reducing hysterectomies. Proposed screening methods include transvaginal ultrasound (TVUS) (not recommended in premenopausal patients) and/or endometrial biopsy. However, current NCCN guidelines suggest that these screening methods may not benefit women with Lynch syndrome. Screening via endometrial biopsy can be considered in patients with Lynch syndrome, due to its high levels of sensitivity and specificity. Screening may begin at age 30 to 35 years and can be repeated every 1 to 2 years. TVUS, on the other hand, is not sensitive or specific at detecting endometrial cancer. However, this screening method can be considered in women with Lynch syndrome based on a provider's judgment.[123]
Two studies have examined the use of TVUS in endometrial screening for women with Lynch syndrome.[482,483] In one study of 292 women from Lynch syndrome families or "Lynch syndrome-like/HNPCC-like" families, TVUS did not detect any endometrial cancer cases. However, two interval cancers developed in symptomatic women.[482] In a second study, 41 women with Lynch syndrome were enrolled in a TVUS screening program. Of 179 TVUS procedures performed, 17 scans were abnormal. Three of the 17 women with abnormal scans had complex atypical hyperplasia on endometrial sampling, while 14 had normal endometrial sampling. However, TVUS failed to identify one patient who presented with abnormal bleeding 8 months after a normal TVUS was performed; this patient was diagnosed with stage IB endometrial cancer.[483] Both of these studies concluded that TVUS is neither sensitive nor specific.
A study of 175 women with Lynch syndrome, which included both endometrial sampling and TVUS, showed that endometrial sampling improved sensitivity when compared with TVUS. Endometrial sampling found 11 of the 14 cases of endometrial cancer. Two of these cases were interval cancers that developed in symptomatic women, and one case was an occult endometrial cancer found at the time of hysterectomy. Endometrial sampling also identified 14 additional cases of endometrial hyperplasia. Among the group of 14 women with endometrial cancer, ten also had TVUS screening with endometrial sampling. Four of the ten women had abnormal TVUS, while six women had normal TVUS.[484] While this cohort study demonstrated that endometrial sampling may have benefits over TVUS for endometrial screening, there are no data that predict that screening with any other modality has benefits for endometrial cancer survival in women with Lynch syndrome.
Some studies suggest that women with a clinical or genetic diagnosis of Lynch syndrome do not participate in intensive gynecologic screening.[485,486] (Refer to the Gynecologic cancer screening for Lynch syndrome section in the Psychosocial Issues in Hereditary Colon Cancer Syndromes section of this summary for more information.)
Ovarian cancer screening in Lynch syndrome
Estimates of the cumulative lifetime risk of ovarian cancer in Lynch syndrome patients range from 3.4% to 22%.[4,365,440-442] NCCN does not recommend routine ovarian cancer screening in individuals with Lynch syndrome. However, CA-125 and pelvic ultrasound are recommended for planning risk-reducing surgery.[123]
Level of evidence: None assigned
Risk-reducing surgeries for the prevention of gynecologic cancers in Lynch syndrome
Risk-reducing surgery is an effective strategy for preventing endometrial and ovarian cancers in Lynch syndrome families. A retrospective study of 315 women with pathogenic MMR gene variants compared the rate of endometrial and ovarian cancers among women who did and did not have hysterectomies and oophorectomies. The mean follow-up periods for endometrial cancer were 13.3 years in the surgical group and 7.4 years in the nonsurgical group. The mean follow-up periods for ovarian cancer were 11.2 years in the surgical group and 10.6 years in the nonsurgical group. In the surgical group, no cancers were diagnosed. In contrast, 33% of women were diagnosed with endometrial cancer, and 5.5% of women were diagnosed with ovarian cancer in the nonsurgical group.[487] Cost-effectiveness–analysis modeling of risk-reducing surgeries (prophylactic hysterectomy and bilateral salpingo-oophorectomy) versus nonsurgical screening was conducted in a theoretical population of 30-year old MMR pathogenic variant carriers. This analysis revealed that prophylactic surgery was cost-effective and yielded higher QALY.[447] A subsequent modeling study evaluated multiple screening and surgical strategies. This study found that annual screening beginning at age 30 years followed by risk-reducing surgery at age 40 years was the most effective strategy.[488]
Additional extracolonic cancer screening in Lynch syndrome
The decision to screen for other Lynch syndrome–associated cancers is done on an individual basis and relies on the cancers reported among FDRs and SDRs with Lynch syndrome.
Gastric and small bowel cancers
The lifetime risk of gastric cancer is approximately 8% for male Lynch syndrome carriers and 5% for female Lynch syndrome carriers.[443] Recent epidemiological data report a decreasing trend in the diagnosis of gastric cancer than was previously reported, which was as high as 13%. The histological characterization of most Lynch syndrome–associated gastric cancers are of the intestinal type and may therefore be detected using screening esophagogastroduodenoscopy (EGD).[443,489] NCCN recommends that EGD with random biopsy of the proximal and distal stomach be initiated at age 30 to 40 years in individuals with Lynch syndrome. This screening can be repeated every 2 to 4 years. Screening can be initiated before age 30 years if any of the following are present: a family history of cancers in the upper GI system, incomplete or extensive gastric intestinal metaplasia, gastric/duodenal adenomas, or Barrett's esophagus with dysplasia. Other risk factors for gastric cancer include the following: male sex, older age, an MLH1 or MSH2 pathogenic variant, Asian ethnicity, residing in or emigrating from countries with a high background incidence of gastric cancer, and chronic autoimmune gastritis.[123]
There are variable reports on the lifetime risk of small bowel cancer associated with Lynch syndrome, ranging from less than 1% to 12%.[4,371,439-441,444] Approximately 6.9% of all duodenal cancer cases are attributed to Lynch syndrome.[79] Most small bowel malignancies are confined to the duodenum and the ileum, which are within endoscopic reach using EGD and colonoscopy (with dedicated ileal intubation), respectively. Other modalities to assess for small bowel lesions include CT enterography and capsule endoscopy but cost-effectiveness analyses do not support use of these evaluations for routine screening in Lynch syndrome.[442]
In a single-center retrospective study, researchers analyzed the prevalence of clinically actionable endoscopic findings in 323 asymptomatic Lynch syndrome carriers undergoing EGD for upper GI cancer surveillance.[490] Sixty-five clinically actionable findings were identified in 57 patients (17.6%), including five individuals with upper GI cancers (1.5%)–which were all detected at an early stage. Cancers were diagnosed at a mean age of 60.2 years (range, 50–73 y). Three of the individuals with upper GI cancers were women. Four out of five individuals with upper GI cancers had an MSH2 pathogenic variant, and one had an MLH1 pathogenic variant. One patient had Barrett's esophagus–related esophageal adenocarcinoma, one patient had a type 3 gastric neuroendocrine tumor, one patient had a gastric adenocarcinoma, and two patients had duodenal adenocarcinomas. These malignancies were detected in Lynch syndrome carriers on baseline EGDs or up to 16 months after surveillance EGDs. The study’s investigators concluded that additional research is needed to investigate the upper GI cancers that developed in the short interval between EGDs.
Esophageal findings in this study included Barrett’s esophagus, which was detected in 6.5% of participants. This percentage is similar to the prevalence of Barrett's esophagus in North American individuals with gastroesophageal reflux disease.[490]
In addition, 261 subjects underwent gastric biopsy sampling, and the following findings were observed:
- Ten patients had H. pylori (3.8%).
- Fifteen patients had gastric intestinal metaplasia (5.7%).
- Five patients had duodenal adenomas (1.5%) at a mean age of 55.8 years (range, 27–75 y). One patient had an adenoma with high-grade dysplasia, and one patient had an adenoma with tubulovillous histology. Four adenomas were found in the third part of the duodenum, and one adenoma was found in the second part of the duodenum.
- Two patients had gastric adenomas (0.6%). One of these patients had an adenoma with high-grade dysplasia. Patients had gastric adenomas at a mean age of 71.6 years (range, 70–73 y).
EGD surveillance is strongly recommended in patients with Lynch syndrome, given the high prevalence and high incidence of clinically actionable findings on EGD and the favorable benefit of identifying these findings at an early stage.
Urinary tract cancer
Urinary tract malignancies include those of the transitional cell type of the renal pelvis and ureters, and the bladder. The associated lifetime risk of these malignancies is variable, ranging from less than 1% to as high as 25%, with higher estimates related to pooling the cancers found in different locations within the urinary tract and including the bladder.[4,371,440,441,444,445] Studies that have evaluated urinary cytology as a potential screening modality revealed that it was associated with low sensitivity and a high false-positive rate and ultimately leads to additional evaluation that is often invasive (i.e., cystoscopy). Screening for renal pelvis, ureter, and/or bladder cancers is generally not recommended in individuals with Lynch syndrome. However, NCCN suggests considering urothelial cancer surveillance for individuals with a family history of urothelial cancer or for individuals (especially men) with an MSH2 pathogenic variant. Annual urinalysis can begin at age 30 to 35 years in these individuals.[123]
Pancreatic cancer
An elevated risk of pancreatic cancer among Lynch syndrome carriers has been supported by two cohort studies that adjust for ascertainment bias. One study reported a cumulative risk of pancreatic cancer of 3.7% by age 70 years and an 8.6-fold increase compared with the general population. [451] Another prospective study using data from the Colon Cancer Family Registry reported an SIR of 10.7 with cumulative risk of 0.95%.[415] Results of these studies have supported an expert consensus that recommended screening for pancreatic cancer in individuals with Lynch syndrome and an FDR with pancreatic cancer, similar to other high-risk populations with comparable risk.[491]
Pancreatic cancer screening may be considered in individuals with MLH1, MSH2, or MSH6 pathogenic variants if they have one or more FDRs or SDRs with exocrine pancreatic cancer (if these family members are on the same side of the family as the individual with Lynch syndrome). Pancreatic cancer screening can begin at age 50 years or 10 years before the youngest pancreatic cancer diagnosis in the family. Screening typically consists of annual contrast-enhanced magnetic resonance imaging/magnetic resonance cholangiopancreatography (MRCP) and/or endoscopic ultrasound (EUS). However, screening can be done more frequently if abnormal findings are found on MRCP/EUS. NCCN recommends that MRCP/EUS occur at a high-volume center that has experience screening individuals with Lynch syndrome. Health care providers are encouraged to have a discussion with patients about pancreatic screening limitations, including the following: the cost of annual pancreatic cancer screening, the high occurrence of benign and indeterminate pancreatic lesions, and the uncertainty regarding the effectiveness of pancreatic cancer screening.[123]
Chemoprevention in Lynch syndrome
The Colorectal Adenoma/Carcinoma Prevention Programme (CAPP2) was a double-blind, placebo-controlled, randomized trial to determine the role of aspirin in preventing CRC in patients with Lynch syndrome who were in surveillance programs at a number of international centers.[492] The study randomly assigned 861 participants (mean age, 45 y) to receive aspirin (600 mg/day), aspirin-placebo, resistant starch (30 g/day), or starch-placebo for up to 4 years. Initial CAPP2 trial results for 746 Lynch syndrome patients enrolled in the study were published in 2008 [493] and failed to show a significant preventive effect on incident colonic adenomas or carcinomas (relative risk, 1.0; 95% CI, 0.7–1.4) with a short-term follow-up (mean, 29 months; range, 7–74 months). In 2011 (mean follow-up, 55.7 months), subsequent data were published on 861 individuals randomly assigned to receive aspirin versus aspirin-placebo, which demonstrated that participants who completed at least 2 years of aspirin (per-protocol analysis) had significantly fewer incident CRCs than individuals randomly assigned to receive aspirin-placebo (HR, 0.41; 95% CI, 0.19–0.86, P = .02; incidence rate ratio [IRR], 0.37; 95% CI, 0.18–0.78, P = .008), although there was no significant difference by intention-to-treat analysis (HR, 0.63; 95% CI, 0.35–1.13, P = .12).[492]
In 2020, long-term follow-up data with all participants having surpassed 10 years of follow-up demonstrated a significant reduction in CRC incidence for participants randomly assigned to receive aspirin both by per-protocol analysis (HR, 0.56; 95% CI, 0.34–0.91) and intention-to-treat analysis (HR, 0.65; 95% CI, 0.43–0.97).[494] These long-term data showed that the protective effects of aspirin did not emerge until approximately 5 years after initiation of aspirin and, intriguingly, the protective effects of aspirin (taken in this study for only a finite period of time, although the data on aspirin use post unblinding do not exist) persisted to at least 20 years of follow-up. These data demonstrated that only 24 Lynch syndrome carriers needed to be treated with this finite course of aspirin to prevent one incident CRC. Female CAPP2 participants randomly assigned to receive aspirin versus aspirin-placebo also developed fewer incident endometrial cancers (7 vs. 17 cases), although this difference did not reach statistical significance (HR, 0.50; 95% CI, 0.22–1.11, P = .09). There was no significant difference seen in the incidence of cancers other than CRC and endometrial cancer in individuals randomly assigned to receive aspirin versus aspirin-placebo. There were no significant differences in adverse events between the aspirin and placebo groups, and no serious adverse effects were noted with any treatment.[492] The CAPP2 investigators concluded that aspirin 600 mg/day has a clear benefit for protecting against CRC in individuals with Lynch syndrome. A key limitation of the CAPP2 trial is that the frequency of surveillance studies at the various centers was not reported as being standardized.
To date, there has been no significant preventive benefit identified in CAPP2 participants randomly assigned to receive resistant starch versus starch-placebo (HR for incident CRC, 0.92; 95% CI, 0.62–1.34; P = .63).[495] However, at a planned 10-year follow-up, participants who had been randomized to receive resistant starch had significantly fewer non-CRC Lynch syndrome cancers than patients who received starch-placebo (IRR, 0.52; 95% CI, 0.32–0.84; P = .0075). This difference seems to have been driven by reduced rates of incident upper-GI tract cancers (gastric, pancreas, biliary, and small bowel cancers).There were 5 upper-GI cancers in participants randomized to take resistant starch and 21 upper-GI cancers in those randomized to take starch-placebo. The biological mechanisms that led to reduced upper-GI tract cancer incidence in Lynch syndrome patients who took resistant starch remain speculative. However, this data have renewed interest in the potential for dietary interventions to be a component of Lynch syndrome cancer prevention.
Experts have speculated that certain Lynch syndrome carriers with lower risks of future incident CRCs (e.g., those with germline PMS2 pathogenic variants, those with prior colectomy, or older individuals) may be less likely to derive benefit from aspirin chemoprevention and may be appropriate for lower dosing.[496] Furthermore, a subgroup analysis of the CAPP2 study also suggested that the chemopreventive effects may be primarily in obese individuals,[497] suggesting that efforts to personalize aspirin chemoprevention recommendations may be appropriate in Lynch syndrome.
The CAPP3 trial, which is evaluating the effect of lower doses of aspirin (blinded 100 mg, 300 mg, and 600 mg enteric-coated aspirin) completed accrual of 1,882 Lynch syndrome carriers in 2019 and data are not expected until at least 5 years of follow-up is complete.[498]
Because of the level 1 evidence in support of aspirin chemoprevention, clinical practice guidelines consistently recommend that individuals with Lynch syndrome consider taking aspirin daily. Optimal aspirin dosage can be determined by the patient's provider, after having a discussion with the patient about his/her personal risk factors (including pregnancy, in which aspirin use may be contraindicated). NCCN also recommends that providers explain potential advantages and disadvantages of aspirin use to the patient.[123]
For Lynch syndrome carriers unable to take aspirin, it is unclear whether NSAIDs may have a comparable chemopreventive benefit. A 2015 survey of 1,858 participants in the Colon Cancer Family Registry suggested that aspirin and ibuprofen might both reduce incident CRC in Lynch syndrome carriers.[499] Additionally, a recent translational study demonstrated that Lynch syndrome carriers randomly assigned to take naproxen for 6 months had significantly lower colorectal mucosal prostaglandin E2 (versus those randomly assigned to take placebo), and that this was accompanied by immune cell activation and gene expression patterns consistent with epithelial differentiation, suggesting the potential underlying mechanism for CRC chemoprevention in Lynch syndrome.[500]
Management of Lynch syndrome-associated CRC
Surgical management of CRC in Lynch syndrome
One of the hallmark features of Lynch syndrome is the presence of synchronous and metachronous CRCs. The incidence of metachronous CRCs has been reported to be 16% at 10 years, 41% at 20 years, and 63% at 30 years after segmental colectomy.[407] Because of the increased incidence of synchronous and metachronous neoplasms, the recommended surgical treatment for a patient with Lynch syndrome with neoplastic colonic lesions is generally an extended colectomy (total or subtotal). Nevertheless, treatment has to be individualized and has often included segmental colectomy. Mathematical models suggest that there are minimal benefits of extended procedures in individuals older than 67 years, compared with the benefits seen in younger individuals with early-onset cancer. In one Markov decision analysis model, the survival advantage for a young individual with early-onset CRC undergoing an extended procedure could be up to 4 years longer than that seen in the same individual undergoing a segmental resection.[501] The recommendation for an extended procedure must be balanced with the comorbidities of the patient, the clinical stage of the disease, the wishes of the patient, and surgical expertise. No prospective or retrospective study has shown a survival advantage for patients with Lynch syndrome who underwent an extended resection versus a segmental procedure.
Two studies have shown that patients who undergo extended procedures have fewer metachronous CRCs and additional surgical procedures related to CRC than do patients who undergo segmental resections.[407,502] Balancing functional results of an extended procedure versus a segmental procedure is of paramount importance. Although most patients adapt well after an abdominal colectomy, some patients will require antidiarrheal medication. A decision model compared QALYs for a patient aged 30 years undergoing an abdominal colectomy versus a segmental colectomy.[503] In this model, there was not much difference between the extended and segmental procedure, with QALYs being 0.3 years more in patients undergoing a segmental procedure than in those undergoing an extended procedure.[503]
A retrospective study from the Creighton University Hereditary Cancer Center evaluated the incidence of metachronous CRC and survival in 64 Lynch syndrome pathogenic variant carriers with right-sided colon cancer undergoing either proximal colectomy or total or subtotal colectomy.[504] Disease incidence was lower in those undergoing a more extensive resection, with one of 16 patients (6.3%) developing metachronous CRC. In those undergoing proximal colectomy, metachronous CRC developed in 13 of 48 patients (27%). There was no statistical difference in survival within a 25-year period after initial surgery. The authors concluded that the treatment of Lynch syndrome patients with right-sided colon cancer at presentation should be individualized to consider quality of life, functional outcomes, and patient preferences.
When considering surgical options, it is important to recognize that a subtotal or total colectomy will not eliminate the rectal cancer risk. The lifetime risk of developing cancer in the rectal remnant after an abdominal colectomy has been reported to be 12% at 12 years post-colectomy.[505] In addition to the general complications of surgery are the potential risks of urinary and sexual dysfunction and diarrhea after an extended colectomy; these risks increase as the anastomosis becomes more distal. Therefore, the choice of surgery must be made on an individual basis by the surgeon and the patient.
In patients with Lynch syndrome and rectal cancer, similar surgical options (extended vs. segmental resection) and considerations must be given. Extended procedures include restorative proctocolectomy and IPAA if the sphincter can be saved, or proctocolectomy with loop ileostomy if the sphincter cannot be saved. The risk of metachronous colon cancer after segmental resection for an index rectal cancer has been reported to be between 15% and 27%.[456,506] Two retrospectives studies reported a 15% and 18% incidence of metachronous colon cancer after segmental rectal cancer–resection in patients with Lynch syndrome.[507,508] In one of the studies, the combined risk of metachronous high-risk adenomas and cancers was 51% at a median follow-up of 101.7 months after proctectomy.[508]
There are no data about fertility after surgery in Lynch syndrome patients. In female FAP patients, no difference in fecundity after abdominal colectomy and IRA has been reported, whereas there is a 54% decrease in fecundity in patients who undergo restorative proctocolectomy with IPAA compared with the general population.[509] Another study in which a questionnaire was sent to FAP patients reported a similar prevalence of fertility problems among patients who had undergone IRA, IPAA, and proctocolectomy with end ileostomy. In that study, it was reported that earlier age at the time of surgery was associated with more fertility problems.[510]
In a large Danish registry study, the incidence rate for metachronous CRC was fivefold higher in Lynch syndrome patients, but not significantly higher in familial CRC and moderate familial risk CRC cases when compared with sporadic CRC, demonstrating that the risk of metachronous CRC occurred almost exclusively in Lynch syndrome cases.[473] The IRRs for metachronous CRC were highest in Lynch syndrome patients with MLH1 and MSH2 germline alterations, intermediate in MSH6 carriers, but not in PMS2 carriers when compared with sporadic CRC cases. Furthermore, when comparing the extent of the colon resections performed for primary CRC in Lynch syndrome cases, the IRRs for metachronous CRC were 5.5 for total and subtotal colectomies and 0.9 for segmental colon resections. The absolute risk reduction of metachronous CRC in Lynch syndrome cases with a primary CRC diagnosed before age 60 years and total/subtotal versus segmental colectomies was 63.5% in men and 40.9% in women. These results strongly support subtotal or total versus segmental colectomies for resectable CRC in Lynch syndrome patients with underlying MLH1 or MSH2, and possibly MSH6 germline alterations.
Most clinicians who treat patients with Lynch syndrome will favor an extended procedure at the time of CRC diagnosis. However, as stated above, the choice of surgery must be made on an individual basis by the surgeon and the patient.[511-513]
Prognostic and therapeutic implications of MSI
As discussed in previous sections, MSI is not only a molecular feature of Lynch syndrome but is also present in 10% to 15% of sporadic cases of CRC (largely due to MLH1 hypermethylation or biallelic somatic variants in an MMR gene). Although MSI testing was initially used to screen patients who might harbor pathogenic MMR gene variants, it has been increasingly recognized that MSI has important prognostic and therapeutic implications. The utility of MSI testing beyond identifying Lynch syndrome has made the case for universal MSI screening more compelling and has contributed to its widespread adoption. Several studies have suggested that stage-specific survival is better for MSI-H CRC compared with MSS cancers. Additionally, the chemotherapeutic agent fluorouracil (5-FU) appears ineffective in the adjuvant treatment of resected MSI-H CRC, in contrast to MSS CRC in which this agent is widely utilized for this purpose. Finally, immunomodulation with agents such as checkpoint inhibitors appears effective in the treatment of advanced MSI-H CRC based on early phase 1 and phase 2 studies, while these agents, at least when utilized as monotherapy, show little activity in MSS CRC.
Prognosis of MSI
Although MSI-H tumors account for 15% of all sporadic CRC, they appear to be more frequent in stage II compared with stage III CRC,[514] and are even less common in metastatic disease, being present in only 3% to 4% of metastatic cases.[515] This stage distinction alludes to the possibility of a better prognosis associated with underlying MSI-H status.
Several studies subsequently confirmed the improved survival of stage II MSI-H CRC compared with MSS cases. A meta-analysis of 32 studies of 7,642 cases, including 1,277 with MSI-H, showed a combined HR estimate for overall survival (OS) associated with MSI of 0.65 (95% CI, 0.59–0.71; heterogeneity P = .16; I2 [a measure of the percentage of variation across studies that is due to heterogeneity rather than chance] = 20%).[516] However, while data were limited, tumors with MSI derived no benefit from adjuvant 5-FU (HR, 1.24; 95% CI, 0.72–2.14). Subsequent data from several large randomized clinical trials confirmed the favorable prognosis associated with MSI-H. These included the QUick And Simple And Reliable (QUASAR) trial, which explored the benefit of adjuvant 5-FU–based chemotherapy compared with surgery alone in 1,900 patients with resected stage II CRC. In this study, MSI-H tumors were associated with a recurrence risk of half that of MSS tumors (risk ratio [RR], 0.53; 95% CI, 0.40–0.70).[517] Similar results were seen in the Pan European Trial Adjuvant Colon Cancer (PETACC)-3 trial, a randomized trial of 5-FU with or without irinotecan in resected stage II or stage III CRC.[518] MSI-H status was associated with an OS odds ratio (OR) of 0.39 (95% CI, 0.24–0.65) and this advantage was seen in both stage II and stage III disease.
Consistent with other prior data, clinicopathologic analysis of 85 Lynch syndrome–associated CRCs and 67 sporadic dMMR CRCs demonstrated a significantly superior survival among patients with Lynch syndrome, as well as younger ages at diagnosis and higher numbers of tumor-infiltrating lymphocytes (TILs).[519] Exome sequencing and neoantigen data from a subset of 16 CRC tumors (eight Lynch syndrome CRCs and eight sporadic dMMR CRCs) from this cohort suggest that somatic mutational burden and neoantigen load is significantly higher among Lynch syndrome–associated CRCs than sporadic dMMR CRCs; this was speculated to be the source of the improved survival outcomes and increased TILs.
Given the predilection for MSI-H tumors to involve the right side of the colon, there is a paucity of data on the outcome and prognosis of MSI-H tumors involving the rectum. One study suggested only 2% of rectal cancers are MSI-H.[517] A study of 62 patients with MSI-H rectal cancers from a single institution were followed for a median of 6.8 years. The 5-year rectal cancer–specific survival was 100% for stage I and stage II, 85.1% for stage III, and 60.0% for stage IV disease, suggesting the favorable prognosis associated with MSI-H may also apply to cancers involving the rectum.[506] The authors additionally reported a favorable 26% pathological complete response rate with 5-FU combined with radiation therapy, suggesting that 5-FU given with radiation for the locoregional treatment of rectal cancer may still be effective in the setting of MSI-H tumors. The substantial rate of pathological complete responses demonstrated in this study also reinforces the need for adequate biopsies to assess MSI status before commencing treatment.
The use of adjuvant chemotherapy after surgery for CRC in Lynch syndrome
The finding of MSI in a CRC has been shown in several studies to predict the lack of benefit of adjuvant chemotherapy with 5-FU in resected stage II or stage III colon cancer.[520] This has been a controversial area historically. It was known that loss of DNA MMR activity in cultured colon cancer cells conferred resistance to DNA-damaging agents (the common mechanism of cytotoxic chemotherapy) through loss of the signal to arrest the cell cycle in response to DNA damage that cannot be repaired.[521] This led to the prediction that DNA dMMR tumors may not be fully sensitive to alkylating agents, 5-FU, and platinum-containing drugs.[522-524] Unexpectedly, in 2000, a paper was published suggesting that patients with Dukes C (stage III) CRC with MSI had a substantial survival benefit when given 5-FU–based adjuvant chemotherapy.[525] However, the patients in this analysis had not been randomized to therapy; they were selected for adjuvant chemotherapy based upon clinical status, and inadvertently, the median age in the treatment group was 13 years younger than the controls.
In 2003, however, the outcomes in a randomized controlled prospective trial of adjuvant chemotherapy in 570 colon cancer patients demonstrated no benefit from adjuvant 5-FU in the group with MSI. Moreover, there were nonsignificant trends towards increased mortality when colon cancers with MSI were treated: twofold for stage III cancers and threefold for stage II cancers.[526] Subsequently, ten studies confirmed this, as all failed to show benefit when CRC patients were given 5-FU–based chemotherapy.[520] In contrast, a meta-analysis of randomized trials of 5-FU versus observation suggested a potential benefit of 5-FU in patients with MSI stage III disease. An exploratory subset analysis suggested benefit only in those patients with Lynch syndrome–related MSI. An analysis of stage II patients was not undertaken in this study.[527]
Preclinical data suggests the addition of oxaliplatin to 5-FU can overcome the resistance to 5-FU monotherapy seen in MSI-H tumors.[528] A retrospective analysis of 433 MSI-H stage II and stage III CRC cases (both sporadic and secondary to Lynch syndrome) suggested a benefit in disease-free survival (DFS) with FOLFOX (5-FU and oxaliplatin) compared with surgery alone.[529] There was a trend towards improved DFS utilizing FOLFOX in the subset of patients with MSI due to Lynch syndrome, however, the result was not statistically significant. Additional studies have demonstrated similar survival outcomes irrespective of MSI status with adjuvant chemotherapy including FOLFOX.[530,531]
Level of evidence (against the use of adjuvant therapy): 1ai
Immunotherapy
Tumors that develop via the MSI pathway have more somatic variants than tumors that develop via other pathways. This could imply that dMMR tumors may have more potential antigens (termed neoantigens) and may be more responsive to immune system manipulation than proficient MMR (pMMR) tumors. Microscopically, MSI-H tumors often exhibit abundant tumor-infiltrating lymphocytes, sometimes resulting in a Crohn-like reaction. This histological feature has long suggested the possibility of increased tumor immune surveillance in MSI-H cancers and is one of the main hypotheses for the better stage-specific survival seen in MSI-H compared with MSS cancers.
To test the hypothesis of efficacy of immunomodulation in MSI-H tumors, a phase 2 trial of programmed cell death-1 (PD-1) inhibition was carried out in a small cohort of patients with MSI-H or MSS cancers. Patients with metastatic disease that had failed various chemotherapy regimens were treated with pembrolizumab, an anti–PD-1 immune checkpoint inhibitor.[532] In this small phase 2 study, 32 patients with CRC (11 were dMMR, 21 were pMMR, and 9 others had noncolorectal dMMR tumors) were treated with intravenous pembrolizumab every 14 days. The immune-related response among evaluable patients was 40% (4 of 10) for dMMR CRC tumors, 0% (0 of 18) for pMMR CRC tumors, and 71% (5 of 7) for non-CRC dMMR tumors. The immune-related 20-week progression-free survival was 78% (7 of 9) in patients with dMMR CRC tumors, 11% (2 of 18) in patients with pMMR CRC tumors, and 67% (4 of 6) in patients with non-CRC dMMR tumors. dMMR tumors had a mean of 24-fold more somatic variants than pMMR tumors. Additionally, in this study somatic variants load was associated with prolonged PFS. The authors concluded that MMR status predicted clinical benefit to immune checkpoint blockade with pembrolizumab.
A single-arm phase 2 study (CheckMate 142) of another PD-1 inhibitor, nivolumab, was performed in 74 patients with MSI-H/dMMR CRC that had progressed on prior cytotoxic chemotherapy (including 5-FU, irinotecan, and oxaliplatin).[533] Overall, 31% of patients (23 of 74) experienced an objective response to therapy, and 69% (51 of 74) had disease control for at least 12 weeks. Among patients who responded to nivolumab, the median duration of response was not reached at the time of study analysis (median follow up of 12 months). There was no significant difference in the response rates among individuals with Lynch syndrome–associated metastatic MSI-H/dMMR CRC versus non-Lynch metastatic MSI-H/dMMR CRC in this study. Twenty percent of study participants experienced grade 3 or greater toxicities, most commonly elevations in amylase and/or lipase, and there were no deaths that were attributed to nivolumab.
Based on these data, pembrolizumab 200 mg given intravenously every 3 weeks was approved by the FDA in May 2017 for the treatment of any MSI-H/dMMR metastatic cancer that is refractory to standard therapy and nivolumab 240 mg given intravenously every 2 weeks was granted accelerated approval by the FDA in August 2017 for the treatment of MSI-H/dMMR CRC that is refractory to cytotoxic chemotherapy.
In 2020, treatment-naïve patients with MSI-H/dMMR CRC were enrolled in a phase III trial (KEYNOTE-177) where they were randomized to receive pembrolizumab or chemotherapy. Patients who received pembrolizumab had an increase in PFS when compared with patients who received chemotherapy.[534,535] Subsequently, pembrolizumab became the standard of care for treating metastatic MSI-H/dMMR CRC in the first-line setting. (Refer to the Colon Cancer Treatment summary for more information.)
In another arm of CheckMate 142, 119 individuals with metastatic dMMR CRC were treated with nivolumab plus ipilimumab.[536] The objective response rate was 55% with a 12-week disease control rate of 80%, a 12-month PFS of 71%, and a median duration of response that was not reached. Grade 3 and grade 4 toxicities occurred in 32% of participants (most commonly increased liver function tests) and 13% of all participants discontinued therapy due to toxicity. This was a nonrandomized study, and thus questions remain as to whether the combination of immune checkpoint blockade is superior to PD-1 inhibition alone, especially given the apparent increase in toxicity with combination therapy. On the basis of these data, in July 2018 the FDA granted accelerated approval to nivolumab plus ipilimumab therapy for the treatment of dMMR/MSI-H metastatic CRC that has progressed through prior chemotherapy with a fluoropyrimidine, oxaliplatin, and irinotecan.
A retrospective analysis described the pathological responses of 14 patients with MSI-H tumors after treatment with PD-1 inhibitors (with or without CTLA-4 inhibitors). Eight of the patients in this study had Lynch syndrome and all of the patients had unresectable/metastatic CRCs. Patients underwent resection after they completed treatment. The study demonstrated a pathological complete response (PCR) in 13 of the 14 patients, despite radiographic evidence of persistent disease in 12 of these patients. The discordance between imaging and PCR may be related to significant lymphocyte infiltration in patient tumors. The median duration of treatment was 12 months. However, a PCR was demonstrated in a patient who was treated for only 3 months.[537]
There is debate about when immunotherapeutic agents can be used in patients with non-CRC, MSI-H cancers. Many providers question in which line of therapy immunotherapeutics should be initiated. This question is the subject of multiple ongoing clinical trials. (Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria.)
Vaccines in the treatment or prevention of MSI-related CRC
An alternative approach to immunotherapy in MSI-H CRC involves the use of tumor-directed vaccines. The most promising approaches thus far involve the use of tumor-related neoantigens as epitopes to increase tumor-specific T-cell immunity. Studies are currently under way in the adjuvant treatment of resected stage III CRC (NCT01461148), in patients with metastatic disease (NCT01885702), and in the prevention of CRC in patients with Lynch syndrome (NCT01885702).
Lynch syndrome–related syndromes
Lynch-like or HNPCC-like syndrome
Somatic biallelic MMR gene inactivation is now recognized as a common cause of sporadic MMR deficiency. The literature variably labels these cases as Lynch-like syndrome (LLS). However, this nomenclature can be confusing, since this is a sporadic form of MMR-deficiency, rather than a stand-alone genetic syndrome. For more information, see the Somatic biallelic mismatch repair deficiency (sometimes called Lynch-like syndrome) section and Table 10.
Hamartomatous Polyposis Syndromes
Hamartomatous polyposis syndromes are a distinct and rare group of polyposis diseases characterized by the presence of hamartomatous growths throughout the GI tract. Each syndrome manifests differently, depending on histological type, the number of GI polyps, the number of benign growths, GI cancer risks, and extra-GI cancer risks. Hamartomatous polyposis syndromes include the following:
PTEN hamartoma tumor syndromes (including Cowden syndrome)
Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome (BRRS) are part of a spectrum of conditions known collectively as PTEN hamartoma tumor syndromes (PHTS). Approximately 85% of patients diagnosed with Cowden syndrome, and approximately 60% of patients with BRRS have an identifiable PTEN pathogenic variant.[538] In addition, PTEN pathogenic variants have been identified in patients with very diverse clinical phenotypes.[539] The term PHTS refers to any patient with a PTEN pathogenic variant, irrespective of clinical presentation.
PTEN functions as a dual-specificity phosphatase that removes phosphate groups from tyrosine, serine, and threonine. PTEN pathogenic variants are diverse and can present as nonsense, missense, frameshift, or splice-site variants. Approximately 40% of variants are found in exon 5, which encodes the phosphatase core motif; several recurrent pathogenic variants have been observed at this location.[540] Pathogenic variants in the 5’ end of PTEN or within the phosphatase core of PTEN tend to affect more organ systems.[541]
Operational criteria for the diagnosis of Cowden syndrome have been published and subsequently updated.[542,543] These include major, minor, and pathognomonic criteria that consist of certain mucocutaneous manifestations and adult-onset dysplastic gangliocytoma of the cerebellum (Lhermitte-Duclos disease). An updated set of criteria based on a systematic literature review has been suggested [544] and is currently utilized in the National Comprehensive Cancer Network (NCCN) guidelines.[545] Contrary to previous criteria, the authors concluded that there was insufficient evidence for any features to be classified as pathognomonic. Increased genetic testing (especially multigene panels) has identified individuals with germline PTEN pathogenic variants who do not meet diagnostic criteria for PHTS. Diagnostic criteria will need to be reconciled with these recently discovered phenotypes. Hence, it is unclear whether PHTS diagnoses should be based on clinical features or a positive PTEN genetic test result. The American College of Medical Genetics and Genomics (ACMG) suggests that referral for genetics consultation be considered for individuals with a personal history of or a first-degree relative with the following: 1) adult-onset Lhermitte-Duclos disease or 2) any three of the major or minor criteria that have been established for the diagnosis of Cowden syndrome.[546] Detailed recommendations, including diagnostic criteria for Cowden syndrome, can be found in the NCCN and ACMG guidelines.[545,546] Additionally, a predictive model that uses clinical criteria to estimate the probability of a PTEN pathogenic variant is available; a cost-effectiveness analysis suggests that germline PTEN testing is cost effective if the probability of a variant is greater than 10%.[547]
Over a 10-year period, the International Cowden Consortium (ICC) prospectively recruited a consecutive series of adult and pediatric patients meeting relaxed ICC criteria for PTEN testing in the United States, Europe, and Asia.[548] Most individuals did not meet the clinical criteria for a diagnosis of Cowden syndrome or BRRS. Of the 3,399 individuals recruited and tested, 295 probands (8.8%) and an additional 73 family members carried a germline PTEN pathogenic variant. The authors concluded that melanoma, kidney cancer, and colorectal cancer should be added to the spectrum of cancers associated with PTEN germline pathogenic variants (in addition to breast cancer, thyroid cancer, and endometrial cancer). This conclusion was based on the high melanoma, kidney, and colorectal cancer lifetime risk estimates found in individuals with PTEN pathogenic variants. A second study of approximately 100 patients with a germline PTEN pathogenic variant confirmed these findings and suggested a cumulative cancer risk of 85% by age 70 years.[549]
The age-adjusted risk of CRC was increased in individuals with PTEN pathogenic variants in both studies (SIR, 5.7–10.3).[548,549] In addition, one study found that 93% of individuals with PTEN pathogenic variants who had undergone at least one colonoscopy had polyps.[548] Hyperplastic polyps were the most common polyp histology identified, although adenomas and sessile serrated polyps were also observed. The increased CRC risk in individuals with PTEN pathogenic variants has led to the recommendation of surveillance colonoscopy in these patients.[549,550] However, both the age at which to begin (30–40 y) and the subsequent frequency of colonoscopies (biennial to every 3–5 y) vary considerably and are based on expert opinion.
| Cancer | Age-Adjusted SIR (95% CI) | Age-Related Penetrance Estimates |
|---|---|---|
| CI = confidence interval; SIR = standardized incidence ratio. | ||
| aAdapted from Tan et al.[548] | ||
| bOther historical studies have suggested a lower lifetime risk of breast cancer, in the range of 25%–50%.[544] (Refer to the PTEN Hamartoma Tumor Syndromes [Including Cowden Syndrome] section in Genetics of Breast and Gynecologic Cancers for more information.) | ||
| Breast | 25.4 (19.8–32.0) | 85% starting around age 30 yb |
| Colorectal | 10.3 (5.6–17.4) | 9% starting around age 40 y |
| Endometrial | 42.9 (28.1–62.8) | 28% starting around age 25 y |
| Kidney | 30.6 (17.8–49.4) | 34% starting around age 40 y |
| Melanoma | 8.5 (4.1–15.6) | 6% with risk beginning in infancy |
| Thyroid | 51.1 (38.1–67.1) | 35% with risk beginning in infancy |
Peutz-Jeghers syndrome
PJS is an early-onset autosomal dominant disorder characterized by melanocytic macules on the lips, the perioral region, and buccal region; and multiple GI polyps, both hamartomatous and adenomatous.[551-553] Germline pathogenic variants in the STK11 gene at chromosome 19p13.3 have been identified in the vast majority of PJS families.[554-558] GI cancers (including colorectal adenocarcinoma, gastric adenocarcinoma, small intestinal adenocarcinoma, and pancreatic adenocarcinoma) are some of the most common malignancies seen in individuals with PJS. PJS also increases the risk of developing cancers in other organs. For example, the cumulative risks have been estimated to be 32% to 54% for breast cancer [8,559,560] and 21% for ovarian cancer (mainly ovarian sex-cord tumors).[559] The risk of developing pancreatic cancer in individuals with PJS is estimated to be more than 100-fold higher than that of the general population (although these statistics are based on calculations from a small number of individuals with PJS).[559] A systematic review found a lifetime cumulative cancer risk, all sites combined, of up to 93% in patients with PJS.[559,561] Table 15 shows the cumulative risk of these tumors.
Females with PJS are also predisposed to the development of cervical adenoma malignum, a rare and very aggressive adenocarcinoma of the cervix.[562] In addition, females with PJS commonly develop benign ovarian sex-cord tumors with annular tubules, whereas males with PJS are predisposed to development of Sertoli-cell testicular tumors;[563] although neither of these two tumor types is malignant, they can cause symptoms related to increased estrogen production.
Although the risk of malignancy appears to be exceedingly high in individuals with PJS based on the published literature, the possibility that selection and referral biases have resulted in overestimates of these risks should be considered.
| Site | Age (y) | Cumulative Risk (%)b | Reference(s) |
|---|---|---|---|
| GI = gastrointestinal. | |||
| aReprinted with permission from Macmillan Publishers Ltd: Gastroenterology [561], copyright 2010. | |||
| bAll cumulative risks were increased compared with the general population (P < .05), with the exception of cervix and testes. | |||
| cGI cancers include colorectal, small intestinal, gastric, esophageal, and pancreatic. | |||
| dWesterman et al.: GI cancer does not include pancreatic cancer.[564] | |||
| eDid not include adenoma malignum of the cervix or Sertoli cell tumors of the testes. | |||
| Any cancer | 60–70 | 37–93 | [8,558-560,564,565] |
| Any GI cancerc,d | 60–70 | 38–66 | [8,560,564,565] |
| Gynecological cancer | 60–70 | 13–18 | [8,560] |
| Per origin | |||
| Stomach | 65 | 29 | [559] |
| Small bowel | 65 | 13 | [559] |
| Colorectum | 65 | 39 | [8,559] |
| Pancreas | 65–70 | 11–36 | [8,559] |
| Lung | 65–70 | 7–17 | [8,559,560] |
| Breast | 60–70 | 32–54 | [8,559,560] |
| Uterus | 65 | 9 | [559] |
| Ovary | 65 | 21 | [559] |
| Cervixe | 65 | 10 | [559] |
| Testese | 65 | 9 | [559] |
PJS is caused by pathogenic variants in the STK11 (also called LKB1) tumor suppressor gene located on chromosome 19p13.[555,556] Unlike the adenomas seen in familial adenomatous polyposis, the polyps arising in PJS are hamartomas. Studies of the hamartomatous polyps and cancers of PJS show allelic imbalance (LOH) consistent with the two-hit hypothesis, demonstrating that STK11 is a tumor suppressor gene.[566,567] However, heterozygous STK11 knockout mice develop hamartomas without inactivation of the remaining wild-type allele, suggesting that haploinsufficiency may be sufficient for initial tumor development in PJS.[568] Subsequently, the cancers that develop in STK11 +/- mice do show LOH;[569] indeed, compound mutant mice heterozygous for pathogenic variants in STK11 +/- and homozygous for pathogenic variants in TP53 -/- have accelerated development of both hamartomas and cancers.[570]
Germline variants of the STK11 gene represent a spectrum of nonsense, frameshift, and missense variants, and splice-site variants and large deletions.[8,554]
Approximately 85% of variants are localized to regions of the kinase domain of the expressed protein. No strong genotype-phenotype correlations have been identified.[8] Up to 30% of variants are large deletions involving one or more exons of STK11, underscoring the importance of deletion analysis in suspected cases of PJS.[554]
STK11 has been unequivocally demonstrated to cause PJS. Although earlier estimates using direct DNA sequencing showed a 50% pathogenic variant detection rate in STK11, studies adding techniques to detect large deletions have found pathogenic variants in up to 94% of individuals meeting clinical criteria for PJS.[554,561,571] Given the results of these studies, it is unlikely that other major genes cause PJS.
Clinical management
NCCN and the U.S. Multi-Society Task Force (USMSTF) on Colorectal Cancer recommend upper endoscopy and high-quality colonoscopy with polypectomy beginning between the ages of 8 to 10 years.[572,573]
Management of small bowel hamartomas is important because patients with PJS have risks of bleeding, intussusception, and malignancy. In PJS, cumulative lifetime risk of small bowel cancer is approximately 13%. NCCN guidelines recommend computed tomography enterography (CTE), magnetic resonance enterography (MRE), or video capsule endoscopy (VCE) beginning between the ages of 8 to 10 years for small bowel surveillance in PJS.[572] These studies are repeated at intervals that are based on study findings up to age 18 years. Afterwards, screening is repeated every 2 to 3 years. Few studies have directly compared yields of these different small bowel cancer surveillance tools. One Australian study of 20 patients with PJS undergoing paired VCE and MRE found that more small bowel polyps (>1 cm) were detected by VCE than MRE.[574] However, balloon enteroscopy detected more small bowel polyps (>1 cm) than both VCE and MRE. NCCN guidelines also include recommendations for other PJS manifestations.
Juvenile polyposis syndrome
JPS is a genetically heterogeneous, rare, childhood- to early adult–onset, autosomal dominant disease that presents characteristically as hamartomatous polyposis throughout the GI tract, although colorectal polyps predominate.[575] JPS can present with diarrhea, GI tract hemorrhage, protein-losing enteropathy, and prolapsing polyps.[575-577] JPS is characterized by the presence of a specific type of hamartomatous polyp called a juvenile polyp, often when a family history of JPS is present. In spite of the histological nomenclature, juvenile polyp diagnosis is based on the polyp's histological appearance, rather than the patient's age when a polyp is found. Solitary juvenile polyps of the colon/rectum are seen sporadically in infants and young children and do not imply a JPS diagnosis.
JPS can be diagnosed clinically (based on fulfillment of specific clinical criteria) or genetically (based on the presence of germline pathogenic variants in SMAD4 or BMPR1A).
A clinical diagnosis of JPS is met by individuals fulfilling one or more of the following criteria:[578]
- More than five juvenile polyps of the colon or rectum.
- Juvenile polyps in other parts of the GI tract.
- Any number of juvenile polyps and a positive family history of JPS.
Approximately 15% to 60% of JPS cases have germline pathogenic variants in the SMAD4 gene, and 25% to 40% of cases have germline pathogenic variants in the BMPR1A gene.[575,579,580]
Individuals with germline SMAD4 pathogenic variants may have a greater risk to develop severe gastric polyposis [581] and hereditary hemorrhagic telangiectasia (HHT) (see the features of HHT below).[575]
Individuals with JPS and germline pathogenic variants in SMAD4/BMPR1A have a 38% to 68% lifetime risk of developing CRC by age 60 years, with an average diagnosis occurring between the ages of 34 and 44 years.[437,582,583] Individuals who have clinical diagnoses of JPS but no identifiable germline SMAD4/BMPR1A pathogenic variants likely have decreased CRC risk. The lifetime risk of gastric cancer in patients with JPS and germline pathogenic variants in SMAD4/BMPR1A has been estimated to be as high as 21%.[123,437] However, the incidences of gastric cancer and gastric polyposis appear to be far lower in individuals with clinical diagnoses of JPS who do not have identifiable germline pathogenic variants in SMAD4/BMPR1A.[583]
Roughly 22% of JPS patients with germline SMAD4 pathogenic variants also have manifestations of HHT, such as arteriovenous malformations, mucocutaneous telangiectasias, digital clubbing, osteoarthropathy, hepatic arteriovenous malformations, and cerebellar cavernous hemangioma, suggesting that the two syndromes overlap.[584] One small case series showed that patients with JPS can infrequently have an adenomatous oligopolyposis phenotype as well.[575,585,586]
NCCN and the USMSTF on Colorectal Cancer recommend that patients with JPS undergo both high-quality colonoscopy with polypectomy and upper endoscopy with polypectomy beginning between ages 12 and 15 years.[123,573] Various clinical practice guidelines recommend that patients with JPS and germline SMAD4 pathogenic variants undergo screening for HHT manifestations.[123,437,575,587]
A severe form of JPS, in which polyposis develops in the first few years of life, is referred to as JPS of infancy. JPS of infancy is often caused by microdeletions of chromosome 10q22-23, a region that includes BMPR1A and PTEN. For more information about PTEN, see the PTEN hamartoma tumor syndromes (including Cowden syndrome) section. The phenotype often includes features such as macrocephaly and developmental delay, possibly as a result of loss of PTEN function.[588] Recurrent GI bleeding, diarrhea, exudative enteropathy, in addition to associated developmental delay, are associated with a very high rate of morbidity and mortality in these infants, thereby limiting the heritability of such cases.[588]
Oligopolyposis
Oligopolyposis (derived from the Greek prefix, oligo, meaning few) describes polyp counts that are greater than anticipated in average-risk patients but are fewer than 100 polyps. Here, oligopolyposis is used to describe situations in which an individual’s polyp count is large enough to suspect a hereditary polyposis syndrome (generally between 10–100 polyps), regardless of the presence or lack of a family history of colorectal polyps/CRC. Since oligopolyposis specifically refers to polyp count, multiple syndromes can present with an oligopolyposis phenotype. It is recommended that most individuals with oligopolyposis have germline genetic testing.[123] These individuals are managed with frequent endoscopic surveillance, based on their underlying hereditary syndrome or guided by their endoscopic presentation.
Multiple syndromes can present with oligopolyposis. These syndromes vary based on polyp histology and the pathogenic variant that is identified. FAP is the most well-described adenomatous polyposis syndrome that occasionally presents with fewer than 100 tubular adenomas. Since the APC gene was discovered in 1992 (pathogenic variants in APC cause FAP), other rare genes have also been implicated in oligopolyposis and cancer. These genes include NTHL1, POLE/POLD1, GREM1, MUTYH, and AXIN2. The hamartomatous polyposis syndromes and serrated polyposis syndrome (SPS) are nonadenomatous polyposis syndromes that can present with oligopolyposis. Individuals with adenomatous polyposis who do not have an identified germline pathogenic variant have colonic adenomatous polyposis of unknown etiology (CPUE).
In a cross-sectional study of 3,789 patients with 10 or more colorectal polyps, patients underwent multigene panel testing for the following genes: APC, BMPR1A, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, PMS2, PTEN, SMAD4, STK11, and TP53. In this study, pathogenic variants were found in 5% of participants, regardless of the participant's age or polyp cohort.[589] GREM1, POLD1, and POLE were only evaluated in 2,353 patients because these genes were added to the multigene panel assay during the study period. The number of pathogenic variants in adenomatous polyposis genes decreased as participants increased in age in all polyp-count groups in the adenoma cohort. This, however, was not true for the nonpolyposis-related CRC genes. The prevalence of pathogenic variants in hamartomatous polyposis genes was high in the hamartoma cohort, regardless of polyp count (40% of participants had 10–19 polyps, 72.1% of participants had 20–99 polyps, and 50% of participants had 100 or more polyps). The study provides additional support for multigene panel testing in individuals with 10 or more colorectal polyps, given that more than 5% of patients had pathogenic variants in all age groups. For more information about genetic testing for polyposis, see the Genetic testing for FAP section.
NTHL1, POLE, POLD1, and GREM1 pathogenic variant testing is being incorporated into the multigene panel tests for CRC susceptibility (offered commercially with APC and MUTYH testing so a polyposis panel can be ordered up front for patients with oligopolyposis). There are minimal data on the optimal surveillance approach for individuals with germline pathogenic variants in NTHL1 (biallelic carriers only), POLE, or POLD1. It is presumed that CRC risk for these pathogenic variants is comparable to that seen in Lynch syndrome. Hence, some guidelines are endorsing early and frequent colonoscopic screening, similar to that which is conducted in Lynch syndrome patients.
Lastly, oligopolyposis with varying polyp histologies (e.g., adenomas, serrated, inflammatory, and hamartomatous polyps) has been reported in two small case series of individuals previously treated with chemotherapy and radiation therapy for a prior childhood malignancy.[590,591] This phenomenon, termed therapy-associated polyposis (TAP), may be an acquired, nonfamilial phenotype caused by prior antineoplastic therapy. TAP is on the differential diagnosis list when nonfamilial oligopolyposis is identified in individuals previously treated with chemotherapy and/or radiation therapy. Another recent study identified oligopolyposis fulfilling World Health Organization (WHO) criteria for serrated polyposis syndrome (SPS) in 6% of a cohort of 101 Hodgkin lymphoma survivors treated with prior chemotherapy and/or radiation therapy.[592] This finding suggests that Hodgkin lymphoma survivors may be a particularly important population in TAP.
Polymerase Proofreading–Associated Polyposis (POLE and POLD1 Genes)
Pathogenic variants in related DNA polymerase genes, POLE and POLD1, have been described in families with oligopolyposis, CRC, and endometrial cancer. This condition is called polymerase proofreading–associated polyposis (PPAP).[593,594] PPAP is an autosomal dominant hereditary cancer syndrome, in which the main replicative DNA polymerases, POLE and POLD1, are unable to proofread newly synthesized DNA strands. Several other malignancies (including endometrial, ovarian, small bowel, and brain cancers), have also been described in POLE and POLD1 carriers. PPAP-associated tumors often present with an ultramutator phenotype characterized by mutational loads exceeding 100 variants per megabase. PPAP-associated cancers can be highly responsive to immune checkpoint inhibitor therapy, irrespective of MSI/MMR status.[595,596] Identifying POLE and POLD1 carriers can be challenging since all known, putative, causal POLE and POLD1 variants are exonuclease-domain missense variants, which are notoriously difficult to classify. In addition, studies on the prevalence and penetrance of PPAP are limited, since this condition is rare, and risk estimates may be subject to ascertainment bias.
The prevalence of POLE and POLD1 pathogenic variants was determined in a prospective cohort of 2,309 unrelated patients with hereditary cancer. These patients all underwent genetic testing with a multigene hereditary cancer panel, in which PPAP was responsible for 0.1% to 0.4% of familial cancer cases (and reaching 0.3% to 0.7% when only CRC and polyposis were considered).[597] The tumors most commonly associated with PPAP included CRC, endometrial cancer, and ovarian cancer.
While PPAP is rare, the age-specific cumulative risks (penetrance) of CRC for POLE and POLD carriers is important to quantify and update, in order to better guide and optimize clinical management. In a retrospective study in which families with POLE or POLD1 pathogenic variants were identified via a PubMed search of relevant studies (prior to 2016) and genotypic data from 669 population-based CRC cases diagnosed in patients under 60 (from the Australasian Colorectal Cancer Family Registry), age-specific cumulative risks were estimated using modified segregation analyses.[598] Sixty-seven CRCs were identified in 364 first-degree relatives (FDRs) and second-degree relatives (SDRs) in 41 POLE families, and 6 CRCs were identified in 69 relatives from 9 POLD1 families. CRC risk to age 70 years for males and females, respectively, was 40% (95% CI, 26%–57%) and 32% (95% CI, 20%–47%) in POLE carriers and 63% (95% CI, 15%–99%) and 52% (95% CI, 11%–99%) in POLD1 carriers. CRC risk for POLE carriers was high enough to consider annual colonoscopy screening and the possible implementation of management guidelines similar to those used in Lynch syndrome. The authors concluded that clinical management recommendations for POLD1 carriers could be similar to those used in POLE carriers, but refinement of POLD1 CRC risk estimates (from larger cohorts) are needed to further refine recommendations.
Lifetime risk estimates of CRC in POLE carriers were higher than expected in a subsequent study of 354 individuals with early-onset/familial CRC (n = 218) and/or attenuated adenomatous polyposis (n = 136) at a single center between 2014 and 2017.[599] In 51 POLE carriers (from 10 families), risk estimates for CRC and extracolonic PPAP-associated cancers were calculated using nonparametric methods (based on a survival analysis approach) to account for unknown genotypes in relatives. Risks of CRC at ages 30 years, 50 years, and 70 years were 11.1% (95% CI, 4.2–17.5), 48.5% (96% CI, 33.2–60.3), and 74% (95% CI, 51.6–86.1), respectively. These risk estimates were similar in both men and women. These results provided additional support for frequent CRC surveillance in POLE carriers, similar to the surveillance used in highly penetrant Lynch syndrome genotypes. Extracolonic cancers like glioblastoma, small bowel cancer, endometrial cancer, and ovarian cancer were observed at low frequencies, limiting the ability to establish screening recommendations for these cancer types.
In POLD1 and POLE carriers, NCCN recommends high-quality colonoscopy starting at age 25 to 30 years. Colonoscopy screening can also begin 2 to 5 years before the earliest CRC in the family, if the CRC was diagnosed before age 25 years. Afterwards, colonoscopy is repeated every 1 to 2 years if polyps are identified or every 2 to 3 years if polyps are not identified.[123] There is insufficient evidence to provide recommendations on extracolonic cancer prevention for patients with variants in these genes.
NTHL1-Associated Polyposis
NTHL1-associated polyposis (or NTHL1 tumor syndrome) was first described in 2015. A study used whole-exome sequencing in 51 individuals (from 48 families) with multiple colonic adenomas.[600] This study found homozygous germline nonsense pathogenic variants in NTHL1 (a base-excision repair gene) in seven affected individuals from three unrelated families. These individuals had CRC and multiple adenomas (8–50 adenomas). None of the adenomas were hyperplastic or serrated. Three affected females also had endometrial cancer or endometrial complex hyperplasia. One individual developed duodenal adenomas, and another individual developed duodenal cancer. All pedigrees were consistent with autosomal recessive inheritance. Three cancers and five adenomas from different affected individuals did not show MSI. These neoplasms did show enrichment of cytosine to thymine transitions.
A subsequent study of 863 families with CRC and 1,600 families without CRC confirmed an association between biallelic NTHL1 pathogenic variants and inherited CRC risk.[601]
In a systematic review of 21 papers, the phenotypic and genotypic spectrum of NTHL1 tumor syndrome, including the occurrence of both benign and malignant tumors, were reported in both biallelic and monoallelic carriers.[602]
- In 47 patients with biallelic pathogenic variants (from 32 families), 23 (49%) were diagnosed with CRC (mean age, 55 y; range, 31–73 y). In addition, 12 of the 22 female patients (55%) were diagnosed with breast cancer (mean age, 49 y; range, 36–63 y). In those who underwent colonoscopy, 93% had colonic adenomas ranging from 2 to 150 polyps, and three patients (6%) had duodenal adenomatosis. Overall, 37 carriers (79%) had at least one primary cancer (cancers occurred in different organs). All ten patients without cancer diagnoses had colonic adenomas. Six female patients (27%) also had gynecological cancers (four patients had endometrial cancer, one patient had ovarian cancer, and one patient had cervical cancer).
- In 158 patients with monoallelic germline NTHL1 variants, 29 individuals (18%) were diagnosed with CRC (mean age, 56 y; range, 36–75 y). Twenty-six out of 68 (38%) monoallelic carriers who underwent colonoscopy had colon polyps or adenomas (1–70 polyps or adenomas). Six patients had more than ten adenomas or polyps. Breast cancer was diagnosed in 59 out of 120 (49%) female monoallelic carriers (mean age, 50 y; range, 24–79 y).
- Limitations of this review included the following: (1) CRC and breast cancer may have been overrepresented due to ascertainment bias; (2) there may have been overlap with other germline pathogenic variants (one Lynch syndrome case and three BRCA2 cases were included in analyses of monoallelic NTHL1 carriers with breast cancer); (3) available data were inconsistent in studies involving monoallelic NTHL1 carriers.
Based on these results, the authors concluded that early endoscopic CRC surveillance can start before age 30 years in biallelic NTHL1 carriers, similar to the surveillance used in MUTYH carriers.[602] Given inconsistent reporting on duodenal polyposis and cancer in biallelic NTHL1 carriers, estimated duodenal neoplasia risk remains unclear, and upper GI tract surveillance could likely be postponed to the third or fourth decade of life. In addition, annual mammography or breast MRI can begin at age 40 years. However, this surveillance can begin at an earlier age if there is a young breast cancer diagnosis in the family. Gynecologic examination with TVUS can be considered every other year, starting at age 40 years.
Consistent with prior reports, monoallelic NTHL1 carriers do not have significantly increased cancer risk. It is not recommended, that these individuals undergo intensive cancer surveillance for GI, breast, or gynecologic cancers.
AXIN2-Related Polyposis
Pathogenic variants in AXIN2, a gene encoding a protein that forms part of the beta-catenin destruction complex, have been associated with oligodontia, ectodermal dysplasia, colorectal polyposis, and CRC in populations enriched with polyposis and CRC.[603,604] The beta-catenin destruction complex consists of several proteins, including AXIN2, and it downregulates beta-catenin, a transcriptional coactivator of Wnt-targeted gene expression.
The prevalence of AXIN2 pathogenic variants and their contribution to both colorectal polyposis and CRC remains unclear. An AXIN2 pathogenic variant was first implicated in a Finnish family (with multiple members) who had oligodontia (absence of >6 nonwisdom teeth in family members), CRC, and polyposis with autosomal dominant inheritance.[605] Subsequent reports had similar results, with other manifestations that included variable penetrance and ectodermal dysplasia (i.e., little hair and eyebrows).[606,607] Affected individuals with AXIN2 pathogenic variants from eight families in Australia/New Zealand were identified based on personal histories of unexplained colonic polyposis (presented with 10 to >100 colorectal polyps) and multiple CRC cases.[603] Similarly, 10 out of 11 French individuals with ten different AXIN2 pathogenic variants were ascertained based on histories of colorectal polyposis or CRC diagnosed at young ages. These individuals presented with adenomatous, serrated, and hyperplastic polyposis by a mean age of 55 years.[604] In three populations, extracolonic cancers included breast cancer, ampullary cancer, ovarian cancer, and melanoma. Multiple cases of tooth agenesis and ectodermal dysplasia (such as sparse hair and eyebrows) were frequently reported.[603,604]
In AXIN2 carriers, NCCN recommends the initiation of high-quality colonoscopy at age 25 to 30 years. Colonoscopy can be repeated every 2 to 3 years if polyps are not identified or every 1 to 2 years if polyps are found. Surgery can be considered in cases with unmanageable polyposis.[123] Given the lack of penetrance data for extracolonic cancers, no further recommendations are provided for AXIN2 carriers.
Hereditary Mixed Polyposis Syndrome (GREM1 Gene)
HMPS is a rare hereditary cancer syndrome characterized by the development of various types of colon polyps including serrated adenomas, atypical juvenile polyps, adenomas, and colonic adenocarcinoma. A small number of individuals/families with HMPS harbor a large, 40-kb, single-copy germline duplication upstream of gremlin 1 (GREM1)—the gene that is thought to cause the HMPS phenotype and increased CRC risk.[608]
Germline GREM1 pathogenic duplications have been observed primarily in individuals/families of Ashkenazi Jewish descent, with varying clinical presentations. Although polyposis appears to be a unifying feature in most families with this duplication, there is a high degree of variability with respect to polyp number, histology, and age of onset. In addition, extracolonic malignancies have been described in several GREM1 pathogenic variant carriers, although the small number of affected individuals limits the ability to definitively demonstrate a causal link to the GREM1 pathogenic variant.
Evidence-based strategies for managing individuals with germline GREM1 pathogenic duplications do not yet exist. However, NCCN guidelines recommend that GREM1 carriers initiate colonoscopic surveillance between ages 25 and 30 years.[123] Colonoscopy is repeated every 2 to 3 years if polyps are not identified or every 1 to 2 years if polyps are identified.
CHEK2
Several studies have suggested that germline CHEK2 pathogenic variants may confer increased risks of colorectal cancer.[609-611] CHEK2 pathogenic variants have been seen in up to 2% of individuals with CRC.[612] However, data have suggested that CHEK2 variants are associated with, at most, a modestly increased risk of CRC (i.e., low penetrance).[613] Two large retrospective studies of CHEK2 cases ascertained from two genetic testing laboratories did not find an increased risk of CRC in CHEK2 carriers.[614,615] In fact, one study found that individuals with germline CHEK2 pathogenic variants had lower risks of CRC when compared with those who had negative results on germline genetic testing panels.[614]
NCCN guidelines recommend that individuals with germline CHEK2 pathogenic variants (and no other CRC risk factors) participate in general population screening for CRC.[123] When a CHEK2 carrier has additional CRC risk factors (e.g., personal/family history of CRC, personal history of colorectal adenomas), the timing and frequency of surveillance colonoscopies is typically determined by the individual's clinical history.
For more information, see the CHEK2 section in Genetics of Breast and Gynecologic Cancers.
Serrated Polyposis Syndrome
The term serrated polyposis syndrome (SPS) has been used to characterize individuals who have mostly sessile serrated polyps/lesions (SSPs/SSLs) in the large intestine. Although the term syndrome is used in SPS, most cases appear to be nonfamilial, and the underlying etiology of these polyps is typically unidentifiable.
Germline evaluation of individuals with serrated polyposis is typically unrevealing.[616-620] Rarely, germline RNF43 pathogenic variants are found in families with serrated polyposis.[621-623] Occasional MUTYH biallelic pathogenic variants and Lynch syndrome pathogenic variants have been found in patients with serrated polyps or features of serrated polyposis.
The WHO states that an individual meets SPS diagnostic criteria if either of the following criteria are fulfilled:[624]
- A personal history of at least five histologically diagnosed serrated lesions (this includes hyperplastic polyps, SSPs, or SSLs) that are all 5 mm or larger, occurring proximal to the rectum. At least two of these polyps/lesions must be 10 mm or larger.
- A personal history of 20 or more serrated lesions (of any size) in the colon/rectum, with 5 or more lesions occurring proximal to the rectum.
Studies have shown that colorectal adenocarcinoma incidence in patients who met formally defined diagnostic criteria for SPS range from 19.9% to greater than 50%.[625-633] A small fraction (<5%) of individuals who fulfill SPS diagnostic criteria harbor germline RNF43 pathogenic variants. Clinical germline genetic testing can be offered to those who meet SPS diagnostic criteria, especially if there is a concomitant family history of CRC and/or SSPs/SSLs.[621-623] While only a small fraction of SPS cases are attributable to monogenic inherited risk, data have suggested that FDRs of individuals with SPS are at increased risk of CRC. Lifestyle factors like smoking, alcohol use, and increased body mass index were associated with SPS in case-control data.[634]
In some individuals with therapy-associated polyposis (TAP), an acquired, mixed-histology form of colorectal and GI tract oligopolyposis, polyps can be seen in childhood/young-adult cancer survivors who were treated with chemotherapy and/or radiotherapy. Some of these individuals will have clinical histories that fulfill SPS diagnostic criteria.[635,636] For more information, see the Oligopolyposis section.
Evidence-based management guidelines for individuals with SPS do not yet exist. NCCN guidelines recommend undergoing high-quality colonoscopy to clear all SSPs/SSLs that are 5 mm or larger (after the SPS phenotype is clinically identified in an individual).[123] Then, colonoscopy is repeated every 1 to 3 years, depending on the number and size of polyps.
MBD4
MBD4-associated neoplasia syndrome is associated with an increased risk of GI polyposis (comparable to AFAP). For more information, see MBD4-Associated Neoplasia Syndrome.
Advances in Endoscopic Imaging in Hereditary CRC
Performance of endoscopic therapies for adenomas in FAP and Lynch syndrome, and decision-making regarding surgical referral and planning, require accurate estimates of the presence of adenomas. In both AFAP and Lynch syndrome the presence of very subtle adenomas poses special challenges—microadenomas in the case of AFAP and flat, though sometimes large, adenomas in Lynch syndrome.
Chromoendoscopy
The need for sensitive means to endoscopically detect subtle polyps has increased with the recognition of flat adenomas and sessile serrated polyps in otherwise average-risk subjects, very attenuated adenoma phenotypes in AFAP, and subtle flat adenomas in Lynch syndrome. Modern high-resolution endoscopes improve adenoma detection yield, but the use of various vital dyes, especially indigo carmine dye-spray, has further improved detection. Several studies have shown that the improved mucosal contrast achieved with the use of indigo carmine can improve the adenoma detection rate. Whether family history is significant or not, careful clinical evaluation consisting of dye-spray colonoscopy (indigo carmine or methylene blue),[474,637-642] with or without magnification, or possibly newer imaging techniques such as narrow-band imaging,[643] may reveal the characteristic right-sided clustering of more numerous microadenomas. Upper GI endoscopy may be informative if duodenal adenomas or fundic gland polyps with surface dysplasia are found. Such findings will increase the likelihood of variant detection if APC or MUTYH testing is pursued.
In various large series of average-risk populations, subtle flat lesions were detected in about 5% to 10% of cases, including adenomas with high-grade dysplasia and invasive adenocarcinoma.[644] Some of these studies involved tandem procedures—white-light exam followed by randomization to “intensive” (> 20-minute pull-back from cecum) inspection versus chromoendoscopy—with significantly more adenomas detected in the chromoendoscopy group.[645] However, in several randomized trials, no significant difference in yield was seen.[646,647]
In a randomized trial of subjects with Lynch syndrome,[648] standard colonoscopy, with polypectomy as indicated, was followed by either indigo carmine chromoendoscopy or repeat “intensive” white-light colonoscopy (a design very nearly identical to the average-risk screening group noted above). In this series, no significant difference in adenoma yield between the chromoendoscopy and intensive white-light groups was detected. However, these patients were younger and, in many cases, had undergone several previous exams that might have resulted in polyp clearing.
In a German study,[649] one series of Lynch syndrome patients underwent white-light exam followed by chromoendoscopy, while a second series underwent colonoscopy with narrow-band imaging followed by chromoendoscopy. Significant differences in flat polyp detection favored chromoendoscopy in both series, although some of the detected lesions were hyperplastic. In a French series of Lynch syndrome subjects that also employed white-light exam followed by chromoendoscopy, significantly more adenomas were detected with chromoendoscopy.[475]
Fewer evaluations of chromoendoscopy have been performed in AFAP than in Lynch syndrome. One study examined four patients with presumed AFAP and fewer than 20 adenomas upon white-light examination.[650] All had more than 1,000 diminutive adenomas found on chromoendoscopy, in agreement with pathology evaluation after colectomy.
A similar role for chromoendoscopy has been suggested to evaluate the duodenum in FAP. One study from Holland that used indigo carmine dye-spray to detect duodenal adenomas showed an increase in the number and size of adenomas, including some large ones. Overall Spigelman score was not significantly affected.[651]
Small bowel imaging
Patients with PJS and JPS are at greater risk of disease-related complications in the small bowel (e.g., bleeding, obstruction, intussusception, or cancer). FAP patients, although at great risk of duodenal neoplasia, have a relatively low risk of jejunoileal involvement. The RR of small bowel malignancy is very high in Lynch syndrome, but absolute risk is less than 10%. Although the risks of small bowel neoplasia are high enough to warrant consideration of surveillance in each disease, the technical challenges of doing so have been daunting. Because of the technical challenges and relatively low prevalences, there is virtually no evidence base for small-bowel screening in Lynch syndrome.
Historically, the relative endoscopic inaccessibility of the mid and distal small bowel required radiographic measures for its evaluation, including the barium small bowel series or a variant called tube enteroclysis, in which a nasogastroduodenal tube is placed so that all of the contrast goes into the small intestine quickly and undiluted by gastric juice for more precise imaging. None of these measures were sensitive for small lesions. Previously, therapeutic removal of lesions required laparotomy. However, multiple novel endoscopic approaches have been developed to overcome the technical limitations of small bowel endoscopy, which has enabled jejunal and ileal access for purposes of polypectomy.
For patients with PJS, double-balloon endoscopy or other forms of deep enteroscopy (single-balloon overtube or spiral overtube) are the preferred methods for evaluation of the small bowel.[652] This may involve either peroral enteroscopy or retrograde enteroscopy to achieve more complete evaluation of the small bowel. Because these procedures are time-consuming and involve risks of complication, deep enteroscopy is usually preceded by more imaging, including barium exams, capsule endoscopy, CT or magnetic resonance enterography.[73]
In FAP, data from capsule endoscopy [73] show a 50% to 100% prevalence of jejunal and/or ileal polyps in patients with Spigelman stage III or stage IV duodenal involvement but virtually no such polyps in Spigelman stage I or stage II disease. Polyps smaller than 10 mm and were not biopsied or removed. Consequently, their clinical significance remains uncertain but is likely limited, given the infrequency of jejunoileal cancer reports in FAP.
Capsule endoscopy in the small series of PJS patients described above [73] showed the presence of a similar frequency (50%–100%) of polyps, but the prevalent polyps were much larger than in FAP, were more likely to become symptomatic, and warranted endoscopic or surgical excision. Capsule studies were suggested as an appropriate replacement for radiographic studies because of the sensitivity of capsule endoscopy.
References
- Bussey HJ: Familial Polyposis Coli: Family Studies, Histopathology, Differential Diagnosis, and Results of Treatment. The Johns Hopkins University Press, 1975.
- Burt RW, Leppert MF, Slattery ML, et al.: Genetic testing and phenotype in a large kindred with attenuated familial adenomatous polyposis. Gastroenterology 127 (2): 444-51, 2004. [PUBMED Abstract]
- Choi YH, Cotterchio M, McKeown-Eyssen G, et al.: Penetrance of colorectal cancer among MLH1/MSH2 carriers participating in the colorectal cancer familial registry in Ontario. Hered Cancer Clin Pract 7 (1): 14, 2009. [PUBMED Abstract]
- Bonadona V, Bonaïti B, Olschwang S, et al.: Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 305 (22): 2304-10, 2011. [PUBMED Abstract]
- Møller P, Seppälä T, Bernstein I, et al.: Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut 66 (3): 464-472, 2017. [PUBMED Abstract]
- Baglietto L, Lindor NM, Dowty JG, et al.: Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 102 (3): 193-201, 2010. [PUBMED Abstract]
- Aretz S, Uhlhaas S, Goergens H, et al.: MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer 119 (4): 807-14, 2006. [PUBMED Abstract]
- Hearle N, Schumacher V, Menko FH, et al.: Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res 12 (10): 3209-15, 2006. [PUBMED Abstract]
- Coburn MC, Pricolo VE, DeLuca FG, et al.: Malignant potential in intestinal juvenile polyposis syndromes. Ann Surg Oncol 2 (5): 386-91, 1995. [PUBMED Abstract]
- Desai DC, Neale KF, Talbot IC, et al.: Juvenile polyposis. Br J Surg 82 (1): 14-7, 1995. [PUBMED Abstract]
- Bülow S, Berk T, Neale K: The history of familial adenomatous polyposis. Fam Cancer 5 (3): 213-20, 2006. [PUBMED Abstract]
- Herrera L, ed.: Familial Adenomatous Polyposis. Alan R. Liss Inc, 1990.
- Bülow S: Familial polyposis coli. Dan Med Bull 34 (1): 1-15, 1987. [PUBMED Abstract]
- Campbell WJ, Spence RA, Parks TG: Familial adenomatous polyposis. Br J Surg 81 (12): 1722-33, 1994. [PUBMED Abstract]
- Inra JA, Steyerberg EW, Grover S, et al.: Racial variation in frequency and phenotypes of APC and MUTYH mutations in 6,169 individuals undergoing genetic testing. Genet Med 17 (10): 815-21, 2015. [PUBMED Abstract]
- Giardiello FM, Offerhaus JG: Phenotype and cancer risk of various polyposis syndromes. Eur J Cancer 31A (7-8): 1085-7, 1995 Jul-Aug. [PUBMED Abstract]
- Jagelman DG, DeCosse JJ, Bussey HJ: Upper gastrointestinal cancer in familial adenomatous polyposis. Lancet 1 (8595): 1149-51, 1988. [PUBMED Abstract]
- Sturt NJ, Gallagher MC, Bassett P, et al.: Evidence for genetic predisposition to desmoid tumours in familial adenomatous polyposis independent of the germline APC mutation. Gut 53 (12): 1832-6, 2004. [PUBMED Abstract]
- Lynch HT, Fitzgibbons R: Surgery, desmoid tumors, and familial adenomatous polyposis: case report and literature review. Am J Gastroenterol 91 (12): 2598-601, 1996. [PUBMED Abstract]
- Bülow S, Björk J, Christensen IJ, et al.: Duodenal adenomatosis in familial adenomatous polyposis. Gut 53 (3): 381-6, 2004. [PUBMED Abstract]
- Burt RW: Colon cancer screening. Gastroenterology 119 (3): 837-53, 2000. [PUBMED Abstract]
- Galiatsatos P, Foulkes WD: Familial adenomatous polyposis. Am J Gastroenterol 101 (2): 385-98, 2006. [PUBMED Abstract]
- Bisgaard ML, Bülow S: Familial adenomatous polyposis (FAP): genotype correlation to FAP phenotype with osteomas and sebaceous cysts. Am J Med Genet A 140 (3): 200-4, 2006. [PUBMED Abstract]
- Petersen GM, Slack J, Nakamura Y: Screening guidelines and premorbid diagnosis of familial adenomatous polyposis using linkage. Gastroenterology 100 (6): 1658-64, 1991. [PUBMED Abstract]
- Berk T, Cohen Z, Bapat B, et al.: Negative genetic test result in familial adenomatous polyposis: clinical screening implications. Dis Colon Rectum 42 (3): 307-10; discussion 310-2, 1999. [PUBMED Abstract]
- Jagelman DG: Clinical management of familial adenomatous polyposis. Cancer Surv 8 (1): 159-67, 1989. [PUBMED Abstract]
- Neale K, Ritchie S, Thomson JP: Screening of offspring of patients with familial adenomatous polyposis: the St. Mark's Hospital polyposis register experience. In: Herrera L, ed.: Familial Adenomatous Polyposis. Alan R. Liss Inc, 1990, pp 61-66.
- Nusliha A, Dalpatadu U, Amarasinghe B, et al.: Congenital hypertrophy of retinal pigment epithelium (CHRPE) in patients with familial adenomatous polyposis (FAP); a polyposis registry experience. BMC Res Notes 7: 734, 2014. [PUBMED Abstract]
- Chen CS, Phillips KD, Grist S, et al.: Congenital hypertrophy of the retinal pigment epithelium (CHRPE) in familial colorectal cancer. Fam Cancer 5 (4): 397-404, 2006. [PUBMED Abstract]
- Coleman P, Barnard NA: Congenital hypertrophy of the retinal pigment epithelium: prevalence and ocular features in the optometric population. Ophthalmic Physiol Opt 27 (6): 547-55, 2007. [PUBMED Abstract]
- Traboulsi EI, Apostolides J, Giardiello FM, et al.: Pigmented ocular fundus lesions and APC mutations in familial adenomatous polyposis. Ophthalmic Genet 17 (4): 167-74, 1996. [PUBMED Abstract]
- Anthony T, Rodriguez-Bigas MA, Weber TK, et al.: Desmoid tumors. J Am Coll Surg 182 (4): 369-77, 1996. [PUBMED Abstract]
- Eccles DM, van der Luijt R, Breukel C, et al.: Hereditary desmoid disease due to a frameshift mutation at codon 1924 of the APC gene. Am J Hum Genet 59 (6): 1193-201, 1996. [PUBMED Abstract]
- Bertario L, Russo A, Sala P, et al.: Genotype and phenotype factors as determinants of desmoid tumors in patients with familial adenomatous polyposis. Int J Cancer 95 (2): 102-7, 2001. [PUBMED Abstract]
- Lynch HT: Desmoid tumors: genotype-phenotype differences in familial adenomatous polyposis--a nosological dilemma. Am J Hum Genet 59 (6): 1184-5, 1996. [PUBMED Abstract]
- Scott RJ, Froggatt NJ, Trembath RC, et al.: Familial infiltrative fibromatosis (desmoid tumours) (MIM135290) caused by a recurrent 3' APC gene mutation. Hum Mol Genet 5 (12): 1921-4, 1996. [PUBMED Abstract]
- Caspari R, Olschwang S, Friedl W, et al.: Familial adenomatous polyposis: desmoid tumours and lack of ophthalmic lesions (CHRPE) associated with APC mutations beyond codon 1444. Hum Mol Genet 4 (3): 337-40, 1995. [PUBMED Abstract]
- Davies DR, Armstrong JG, Thakker N, et al.: Severe Gardner syndrome in families with mutations restricted to a specific region of the APC gene. Am J Hum Genet 57 (5): 1151-8, 1995. [PUBMED Abstract]
- Bertario L, Russo A, Sala P, et al.: Multiple approach to the exploration of genotype-phenotype correlations in familial adenomatous polyposis. J Clin Oncol 21 (9): 1698-707, 2003. [PUBMED Abstract]
- Elayi E, Manilich E, Church J: Polishing the crystal ball: knowing genotype improves ability to predict desmoid disease in patients with familial adenomatous polyposis. Dis Colon Rectum 52 (10): 1762-6, 2009. [PUBMED Abstract]
- Nieuwenhuis MH, Lefevre JH, Bülow S, et al.: Family history, surgery, and APC mutation are risk factors for desmoid tumors in familial adenomatous polyposis: an international cohort study. Dis Colon Rectum 54 (10): 1229-34, 2011. [PUBMED Abstract]
- Clark SK, Smith TG, Katz DE, et al.: Identification and progression of a desmoid precursor lesion in patients with familial adenomatous polyposis. Br J Surg 85 (7): 970-3, 1998. [PUBMED Abstract]
- Hodgson SV, Maher ER: Gastro-intestinal system. In: Hodgson SV, Maher ER: A Practical Guide to Human Cancer Genetics. 2nd ed. Cambridge University Press, 1999, pp 167-175.
- Rodriguez-Bigas MA, Mahoney MC, Karakousis CP, et al.: Desmoid tumors in patients with familial adenomatous polyposis. Cancer 74 (4): 1270-4, 1994. [PUBMED Abstract]
- Clark SK, Neale KF, Landgrebe JC, et al.: Desmoid tumours complicating familial adenomatous polyposis. Br J Surg 86 (9): 1185-9, 1999. [PUBMED Abstract]
- Belchetz LA, Berk T, Bapat BV, et al.: Changing causes of mortality in patients with familial adenomatous polyposis. Dis Colon Rectum 39 (4): 384-7, 1996. [PUBMED Abstract]
- Iwama T, Tamura K, Morita T, et al.: A clinical overview of familial adenomatous polyposis derived from the database of the Polyposis Registry of Japan. Int J Clin Oncol 9 (4): 308-16, 2004. [PUBMED Abstract]
- Church J, Berk T, Boman BM, et al.: Staging intra-abdominal desmoid tumors in familial adenomatous polyposis: a search for a uniform approach to a troubling disease. Dis Colon Rectum 48 (8): 1528-34, 2005. [PUBMED Abstract]
- Parc Y, Piquard A, Dozois RR, et al.: Long-term outcome of familial adenomatous polyposis patients after restorative coloproctectomy. Ann Surg 239 (3): 378-82, 2004. [PUBMED Abstract]
- Church JM, McGannon E, Hull-Boiner S, et al.: Gastroduodenal polyps in patients with familial adenomatous polyposis. Dis Colon Rectum 35 (12): 1170-3, 1992. [PUBMED Abstract]
- Sarre RG, Frost AG, Jagelman DG, et al.: Gastric and duodenal polyps in familial adenomatous polyposis: a prospective study of the nature and prevalence of upper gastrointestinal polyps. Gut 28 (3): 306-14, 1987. [PUBMED Abstract]
- Watanabe H, Enjoji M, Yao T, et al.: Gastric lesions in familial adenomatosis coli: their incidence and histologic analysis. Hum Pathol 9 (3): 269-83, 1978. [PUBMED Abstract]
- Weston BR, Helper DJ, Rex DK: Positive predictive value of endoscopic features deemed typical of gastric fundic gland polyps. J Clin Gastroenterol 36 (5): 399-402, 2003 May-Jun. [PUBMED Abstract]
- Abraham SC, Nobukawa B, Giardiello FM, et al.: Fundic gland polyps in familial adenomatous polyposis: neoplasms with frequent somatic adenomatous polyposis coli gene alterations. Am J Pathol 157 (3): 747-54, 2000. [PUBMED Abstract]
- Odze RD, Marcial MA, Antonioli D: Gastric fundic gland polyps: a morphological study including mucin histochemistry, stereometry, and MIB-1 immunohistochemistry. Hum Pathol 27 (9): 896-903, 1996. [PUBMED Abstract]
- Wu TT, Kornacki S, Rashid A, et al.: Dysplasia and dysregulation of proliferation in foveolar and surface epithelia of fundic gland polyps from patients with familial adenomatous polyposis. Am J Surg Pathol 22 (3): 293-8, 1998. [PUBMED Abstract]
- Burt RW: Gastric fundic gland polyps. Gastroenterology 125 (5): 1462-9, 2003. [PUBMED Abstract]
- Bianchi LK, Burke CA, Bennett AE, et al.: Fundic gland polyp dysplasia is common in familial adenomatous polyposis. Clin Gastroenterol Hepatol 6 (2): 180-5, 2008. [PUBMED Abstract]
- Jalving M, Koornstra JJ, Wesseling J, et al.: Increased risk of fundic gland polyps during long-term proton pump inhibitor therapy. Aliment Pharmacol Ther 24 (9): 1341-8, 2006. [PUBMED Abstract]
- Leggett B: FAP: another indication to treat H pylori. Gut 51 (4): 463-4, 2002. [PUBMED Abstract]
- Nakamura S, Matsumoto T, Kobori Y, et al.: Impact of Helicobacter pylori infection and mucosal atrophy on gastric lesions in patients with familial adenomatous polyposis. Gut 51 (4): 485-9, 2002. [PUBMED Abstract]
- Iida M, Yao T, Itoh H, et al.: Natural history of gastric adenomas in patients with familial adenomatosis coli/Gardner's syndrome. Cancer 61 (3): 605-11, 1988. [PUBMED Abstract]
- Bülow S, Alm T, Fausa O, et al.: Duodenal adenomatosis in familial adenomatous polyposis. DAF Project Group. Int J Colorectal Dis 10 (1): 43-6, 1995. [PUBMED Abstract]
- Park JG, Park KJ, Ahn YO, et al.: Risk of gastric cancer among Korean familial adenomatous polyposis patients. Report of three cases. Dis Colon Rectum 35 (10): 996-8, 1992. [PUBMED Abstract]
- Iwama T, Mishima Y, Utsunomiya J: The impact of familial adenomatous polyposis on the tumorigenesis and mortality at the several organs. Its rational treatment. Ann Surg 217 (2): 101-8, 1993. [PUBMED Abstract]
- Offerhaus GJ, Giardiello FM, Krush AJ, et al.: The risk of upper gastrointestinal cancer in familial adenomatous polyposis. Gastroenterology 102 (6): 1980-2, 1992. [PUBMED Abstract]
- Brosens LA, Keller JJ, Offerhaus GJ, et al.: Prevention and management of duodenal polyps in familial adenomatous polyposis. Gut 54 (7): 1034-43, 2005. [PUBMED Abstract]
- Ngamruengphong S, Boardman LA, Heigh RI, et al.: Gastric adenomas in familial adenomatous polyposis are common, but subtle, and have a benign course. Hered Cancer Clin Pract 12 (1): 4, 2014. [PUBMED Abstract]
- Mankaney G, Leone P, Cruise M, et al.: Gastric cancer in FAP: a concerning rise in incidence. Fam Cancer 16 (3): 371-376, 2017. [PUBMED Abstract]
- Li J, Woods SL, Healey S, et al.: Point Mutations in Exon 1B of APC Reveal Gastric Adenocarcinoma and Proximal Polyposis of the Stomach as a Familial Adenomatous Polyposis Variant. Am J Hum Genet 98 (5): 830-842, 2016. [PUBMED Abstract]
- Perzin KH, Bridge MF: Adenomas of the small intestine: a clinicopathologic review of 51 cases and a study of their relationship to carcinoma. Cancer 48 (3): 799-819, 1981. [PUBMED Abstract]
- Ranzi T, Castagnone D, Velio P, et al.: Gastric and duodenal polyps in familial polyposis coli. Gut 22 (5): 363-7, 1981. [PUBMED Abstract]
- Burke CA, Santisi J, Church J, et al.: The utility of capsule endoscopy small bowel surveillance in patients with polyposis. Am J Gastroenterol 100 (7): 1498-502, 2005. [PUBMED Abstract]
- Tescher P, Macrae FA, Speer T, et al.: Surveillance of FAP: a prospective blinded comparison of capsule endoscopy and other GI imaging to detect small bowel polyps. Hered Cancer Clin Pract 8 (1): 3, 2010. [PUBMED Abstract]
- Eliakim R: Video capsule endoscopy of the small bowel. Curr Opin Gastroenterol 26 (2): 129-33, 2010. [PUBMED Abstract]
- Taylor SA, Halligan S, Moore L, et al.: Multidetector-row CT duodenography in familial adenomatous polyposis: a pilot study. Clin Radiol 59 (10): 939-45, 2004. [PUBMED Abstract]
- Vasen HF, Bülow S, Myrhøj T, et al.: Decision analysis in the management of duodenal adenomatosis in familial adenomatous polyposis. Gut 40 (6): 716-9, 1997. [PUBMED Abstract]
- Groves CJ, Saunders BP, Spigelman AD, et al.: Duodenal cancer in patients with familial adenomatous polyposis (FAP): results of a 10 year prospective study. Gut 50 (5): 636-41, 2002. [PUBMED Abstract]
- Aparicio T, Henriques J, Manfredi S, et al.: Small bowel adenocarcinoma: Results from a nationwide prospective ARCAD-NADEGE cohort study of 347 patients. Int J Cancer 147 (4): 967-977, 2020. [PUBMED Abstract]
- Bleau BL, Gostout CJ: Endoscopic treatment of ampullary adenomas in familial adenomatous polyposis. J Clin Gastroenterol 22 (3): 237-41, 1996. [PUBMED Abstract]
- Norton ID, Gostout CJ: Management of periampullary adenoma. Dig Dis 16 (5): 266-73, 1998 Sep-Oct. [PUBMED Abstract]
- Norton ID, Gostout CJ, Baron TH, et al.: Safety and outcome of endoscopic snare excision of the major duodenal papilla. Gastrointest Endosc 56 (2): 239-43, 2002. [PUBMED Abstract]
- Saurin JC, Gutknecht C, Napoleon B, et al.: Surveillance of duodenal adenomas in familial adenomatous polyposis reveals high cumulative risk of advanced disease. J Clin Oncol 22 (3): 493-8, 2004. [PUBMED Abstract]
- Spigelman AD, Williams CB, Talbot IC, et al.: Upper gastrointestinal cancer in patients with familial adenomatous polyposis. Lancet 2 (8666): 783-5, 1989. [PUBMED Abstract]
- Thiruvengadam SS, Lopez R, O'Malley M, et al.: Spigelman stage IV duodenal polyposis does not precede most duodenal cancer cases in patients with familial adenomatous polyposis. Gastrointest Endosc 89 (2): 345-354.e2, 2019. [PUBMED Abstract]
- Cetta F, Montalto G, Gori M, et al.: Germline mutations of the APC gene in patients with familial adenomatous polyposis-associated thyroid carcinoma: results from a European cooperative study. J Clin Endocrinol Metab 85 (1): 286-92, 2000. [PUBMED Abstract]
- Cetta F, Curia MC, Montalto G, et al.: Thyroid carcinoma usually occurs in patients with familial adenomatous polyposis in the absence of biallelic inactivation of the adenomatous polyposis coli gene. J Clin Endocrinol Metab 86 (1): 427-32, 2001. [PUBMED Abstract]
- Seki M, Tanaka K, Kikuchi-Yanoshita R, et al.: Loss of normal allele of the APC gene in an adrenocortical carcinoma from a patient with familial adenomatous polyposis. Hum Genet 89 (3): 298-300, 1992. [PUBMED Abstract]
- Marchesa P, Fazio VW, Church JM, et al.: Adrenal masses in patients with familial adenomatous polyposis. Dis Colon Rectum 40 (9): 1023-8, 1997. [PUBMED Abstract]
- Kallenberg FGJ, Bastiaansen BAJ, Nio CY, et al.: Adrenal Lesions in Patients With (Attenuated) Familial Adenomatous Polyposis and MUTYH-Associated Polyposis. Dis Colon Rectum 60 (10): 1057-1064, 2017. [PUBMED Abstract]
- Cetta F, Mazzarella L, Bon G, et al.: Genetic alterations in hepatoblastoma and hepatocellular carcinoma associated with familial adenomatous polyposis. Med Pediatr Oncol 41 (5): 496-7, 2003. [PUBMED Abstract]
- Young J, Barker M, Robertson T, et al.: A case of myoepithelial carcinoma displaying biallelic inactivation of the tumour suppressor gene APC in a patient with familial adenomatous polyposis. J Clin Pathol 55 (3): 230-1, 2002. [PUBMED Abstract]
- Cetta F, Montalto G, Petracci M: Hepatoblastoma and APC gene mutation in familial adenomatous polyposis. Gut 41 (3): 417, 1997. [PUBMED Abstract]
- Giardiello FM, Petersen GM, Brensinger JD, et al.: Hepatoblastoma and APC gene mutation in familial adenomatous polyposis. Gut 39 (96): 867-9, 1996. [PUBMED Abstract]
- Ding SF, Michail NE, Habib NA: Genetic changes in hepatoblastoma. J Hepatol 20 (5): 672-5, 1994. [PUBMED Abstract]
- Hughes LJ, Michels VV: Risk of hepatoblastoma in familial adenomatous polyposis. Am J Med Genet 43 (6): 1023-5, 1992. [PUBMED Abstract]
- Bernstein IT, Bülow S, Mauritzen K: Hepatoblastoma in two cousins in a family with adenomatous polyposis. Report of two cases. Dis Colon Rectum 35 (4): 373-4, 1992. [PUBMED Abstract]
- Giardiello FM, Offerhaus GJ, Krush AJ, et al.: Risk of hepatoblastoma in familial adenomatous polyposis. J Pediatr 119 (5): 766-8, 1991. [PUBMED Abstract]
- Perilongo G: Link confirmed between FAP and hepatoblastoma. Oncology (Huntingt) 5 (7): 14, 1991. [PUBMED Abstract]
- Toyama WM, Wagner S: Hepatoblastoma with familial polyposis coli: another case and corrected pedigree. Surgery 108 (1): 123-4, 1990. [PUBMED Abstract]
- Kurahashi H, Takami K, Oue T, et al.: Biallelic inactivation of the APC gene in hepatoblastoma. Cancer Res 55 (21): 5007-11, 1995. [PUBMED Abstract]
- Hirschman BA, Pollock BH, Tomlinson GE: The spectrum of APC mutations in children with hepatoblastoma from familial adenomatous polyposis kindreds. J Pediatr 147 (2): 263-6, 2005. [PUBMED Abstract]
- Hamilton SR, Liu B, Parsons RE, et al.: The molecular basis of Turcot's syndrome. N Engl J Med 332 (13): 839-47, 1995. [PUBMED Abstract]
- Laurent-Puig P, Béroud C, Soussi T: APC gene: database of germline and somatic mutations in human tumors and cell lines. Nucleic Acids Res 26 (1): 269-70, 1998. [PUBMED Abstract]
- Yan H, Dobbie Z, Gruber SB, et al.: Small changes in expression affect predisposition to tumorigenesis. Nat Genet 30 (1): 25-6, 2002. [PUBMED Abstract]
- Spirio L, Olschwang S, Groden J, et al.: Alleles of the APC gene: an attenuated form of familial polyposis. Cell 75 (5): 951-7, 1993. [PUBMED Abstract]
- Brensinger JD, Laken SJ, Luce MC, et al.: Variable phenotype of familial adenomatous polyposis in pedigrees with 3' mutation in the APC gene. Gut 43 (4): 548-52, 1998. [PUBMED Abstract]
- Soravia C, Berk T, Madlensky L, et al.: Genotype-phenotype correlations in attenuated adenomatous polyposis coli. Am J Hum Genet 62 (6): 1290-301, 1998. [PUBMED Abstract]
- Pedemonte S, Sciallero S, Gismondi V, et al.: Novel germline APC variants in patients with multiple adenomas. Genes Chromosomes Cancer 22 (4): 257-67, 1998. [PUBMED Abstract]
- Rozen P, Samuel Z, Shomrat R, et al.: Notable intrafamilial phenotypic variability in a kindred with familial adenomatous polyposis and an APC mutation in exon 9. Gut 45 (6): 829-33, 1999. [PUBMED Abstract]
- Fearnhead NS: Familial adenomatous polyposis and MYH. Lancet 362 (9377): 5-6, 2003. [PUBMED Abstract]
- Al-Tassan N, Chmiel NH, Maynard J, et al.: Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet 30 (2): 227-32, 2002. [PUBMED Abstract]
- Bellido F, Pineda M, Aiza G, et al.: POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med 18 (4): 325-32, 2016. [PUBMED Abstract]
- Spier I, Holzapfel S, Altmüller J, et al.: Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int J Cancer 137 (2): 320-31, 2015. [PUBMED Abstract]
- Bakry D, Aronson M, Durno C, et al.: Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur J Cancer 50 (5): 987-96, 2014. [PUBMED Abstract]
- Grover S, Kastrinos F, Steyerberg EW, et al.: Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA 308 (5): 485-92, 2012. [PUBMED Abstract]
- Sieber OM, Lamlum H, Crabtree MD, et al.: Whole-gene APC deletions cause classical familial adenomatous polyposis, but not attenuated polyposis or "multiple" colorectal adenomas. Proc Natl Acad Sci U S A 99 (5): 2954-8, 2002. [PUBMED Abstract]
- Michils G, Tejpar S, Thoelen R, et al.: Large deletions of the APC gene in 15% of mutation-negative patients with classical polyposis (FAP): a Belgian study. Hum Mutat 25 (2): 125-34, 2005. [PUBMED Abstract]
- Kadiyska TK, Todorov TP, Bichev SN, et al.: APC promoter 1B deletion in familial polyposis--implications for mutation-negative families. Clin Genet 85 (5): 452-7, 2014. [PUBMED Abstract]
- Jasperson KW, Patel SG, Ahnen DJ, et al., eds.: APC-Associated Polyposis Conditions. In: Adam MP, Feldman J, Mirzaa GM, et al., eds.: GeneReviews. University of Washington, Seattle, 1993-2024, pp. Available online. Last accessed May 24, 2024.
- Bisgaard ML, Fenger K, Bülow S, et al.: Familial adenomatous polyposis (FAP): frequency, penetrance, and mutation rate. Hum Mutat 3 (2): 121-5, 1994. [PUBMED Abstract]
- Patenaude AF: Cancer susceptibility testing: risks, benefits, and personal beliefs. In: Clarke A, ed.: The Genetic Testing of Children. BIOS Scientific, 1998, pp 145-156.
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal, Endometrial, and Gastric. Version 3.2024. Plymouth Meeting, PA: National Comprehensive Cancer Network, 2024. Available with free registration. Last accessed December 13, 2024.
- Nugent KP, Spigelman AD, Phillips RK: Life expectancy after colectomy and ileorectal anastomosis for familial adenomatous polyposis. Dis Colon Rectum 36 (11): 1059-62, 1993. [PUBMED Abstract]
- Barrow P, Khan M, Lalloo F, et al.: Systematic review of the impact of registration and screening on colorectal cancer incidence and mortality in familial adenomatous polyposis and Lynch syndrome. Br J Surg 100 (13): 1719-31, 2013. [PUBMED Abstract]
- Koskenvuo L, Pitkäniemi J, Rantanen M, et al.: Impact of Screening on Survival in Familial Adenomatous Polyposis. J Clin Gastroenterol 50 (1): 40-4, 2016. [PUBMED Abstract]
- Hakulinen T, Seppä K, Lambert PC: Choosing the relative survival method for cancer survival estimation. Eur J Cancer 47 (14): 2202-10, 2011. [PUBMED Abstract]
- Petersen GM: Genetic testing and counseling in familial adenomatous polyposis. Oncology (Huntingt) 10 (1): 89-94; discussion 97-8, 1996. [PUBMED Abstract]
- Church J, Burke C, McGannon E, et al.: Risk of rectal cancer in patients after colectomy and ileorectal anastomosis for familial adenomatous polyposis: a function of available surgical options. Dis Colon Rectum 46 (9): 1175-81, 2003. [PUBMED Abstract]
- Guillem JG, Wood WC, Moley JF, et al.: ASCO/SSO review of current role of risk-reducing surgery in common hereditary cancer syndromes. Ann Surg Oncol 13 (10): 1296-321, 2006. [PUBMED Abstract]
- Bertario L, Russo A, Radice P, et al.: Genotype and phenotype factors as determinants for rectal stump cancer in patients with familial adenomatous polyposis. Hereditary Colorectal Tumors Registry. Ann Surg 231 (4): 538-43, 2000. [PUBMED Abstract]
- Heiskanen I, Järvinen HJ: Fate of the rectal stump after colectomy and ileorectal anastomosis for familial adenomatous polyposis. Int J Colorectal Dis 12 (1): 9-13, 1997. [PUBMED Abstract]
- Bülow S, Bülow C, Vasen H, et al.: Colectomy and ileorectal anastomosis is still an option for selected patients with familial adenomatous polyposis. Dis Colon Rectum 51 (9): 1318-23, 2008. [PUBMED Abstract]
- De Cosse JJ, Bülow S, Neale K, et al.: Rectal cancer risk in patients treated for familial adenomatous polyposis. The Leeds Castle Polyposis Group. Br J Surg 79 (12): 1372-5, 1992. [PUBMED Abstract]
- Bess MA, Adson MA, Elveback LR, et al.: Rectal cancer following colectomy for polyposis. Arch Surg 115 (4): 460-7, 1980. [PUBMED Abstract]
- Iwama T, Mishima Y: Factors affecting the risk of rectal cancer following rectum-preserving surgery in patients with familial adenomatous polyposis. Dis Colon Rectum 37 (10): 1024-6, 1994. [PUBMED Abstract]
- Bülow S, Højen H, Buntzen S, et al.: Primary and secondary restorative proctocolectomy for familial adenomatous polyposis: complications and long-term bowel function. Colorectal Dis 15 (4): 436-41, 2013. [PUBMED Abstract]
- Church J, Burke C, McGannon E, et al.: Predicting polyposis severity by proctoscopy: how reliable is it? Dis Colon Rectum 44 (9): 1249-54, 2001. [PUBMED Abstract]
- Nieuwenhuis MH, Bülow S, Björk J, et al.: Genotype predicting phenotype in familial adenomatous polyposis: a practical application to the choice of surgery. Dis Colon Rectum 52 (7): 1259-63, 2009. [PUBMED Abstract]
- Vasen HF, van der Luijt RB, Slors JF, et al.: Molecular genetic tests as a guide to surgical management of familial adenomatous polyposis. Lancet 348 (9025): 433-5, 1996. [PUBMED Abstract]
- Wu JS, Paul P, McGannon EA, et al.: APC genotype, polyp number, and surgical options in familial adenomatous polyposis. Ann Surg 227 (1): 57-62, 1998. [PUBMED Abstract]
- Nieuwenhuis MH, Mathus-Vliegen LM, Slors FJ, et al.: Genotype-phenotype correlations as a guide in the management of familial adenomatous polyposis. Clin Gastroenterol Hepatol 5 (3): 374-8, 2007. [PUBMED Abstract]
- Lovegrove RE, Tilney HS, Heriot AG, et al.: A comparison of adverse events and functional outcomes after restorative proctocolectomy for familial adenomatous polyposis and ulcerative colitis. Dis Colon Rectum 49 (9): 1293-306, 2006. [PUBMED Abstract]
- Parc YR, Olschwang S, Desaint B, et al.: Familial adenomatous polyposis: prevalence of adenomas in the ileal pouch after restorative proctocolectomy. Ann Surg 233 (3): 360-4, 2001. [PUBMED Abstract]
- Groves CJ, Beveridge G, Swain DJ, et al.: Prevalence and morphology of pouch and ileal adenomas in familial adenomatous polyposis. Dis Colon Rectum 48 (4): 816-23, 2005. [PUBMED Abstract]
- Lee CHA, Kalady MF, Burke CA, et al.: Incidence and Management of Rectal Cuff and Anal Transitional Zone Neoplasia in Patients With Familial Adenomatous Polyposis. Dis Colon Rectum 64 (8): 977-985, 2021. [PUBMED Abstract]
- Ooi BS, Remzi FH, Gramlich T, et al.: Anal transitional zone cancer after restorative proctocolectomy and ileoanal anastomosis in familial adenomatous polyposis: report of two cases. Dis Colon Rectum 46 (10): 1418-23; discussion 1422-3, 2003. [PUBMED Abstract]
- Steinbach G, Lynch PM, Phillips RK, et al.: The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N Engl J Med 342 (26): 1946-52, 2000. [PUBMED Abstract]
- Giardiello FM, Yang VW, Hylind LM, et al.: Primary chemoprevention of familial adenomatous polyposis with sulindac. N Engl J Med 346 (14): 1054-9, 2002. [PUBMED Abstract]
- Lynch PM, Burke CA, Phillips R, et al.: An international randomised trial of celecoxib versus celecoxib plus difluoromethylornithine in patients with familial adenomatous polyposis. Gut 65 (2): 286-95, 2016. [PUBMED Abstract]
- Burke CA, Dekker E, Lynch P, et al.: Eflornithine plus Sulindac for Prevention of Progression in Familial Adenomatous Polyposis. N Engl J Med 383 (11): 1028-1039, 2020. [PUBMED Abstract]
- Balaguer F, Stoffel EM, Burke CA, et al.: Combination of Sulindac and Eflornithine Delays the Need for Lower Gastrointestinal Surgery in Patients With Familial Adenomatous Polyposis: Post Hoc Analysis of a Randomized Clinical Trial. Dis Colon Rectum 65 (4): 536-545, 2022. [PUBMED Abstract]
- Lynch PM, Ayers GD, Hawk E, et al.: The safety and efficacy of celecoxib in children with familial adenomatous polyposis. Am J Gastroenterol 105 (6): 1437-43, 2010. [PUBMED Abstract]
- West NJ, Clark SK, Phillips RK, et al.: Eicosapentaenoic acid reduces rectal polyp number and size in familial adenomatous polyposis. Gut 59 (7): 918-25, 2010. [PUBMED Abstract]
- Phillips RK, Wallace MH, Lynch PM, et al.: A randomised, double blind, placebo controlled study of celecoxib, a selective cyclooxygenase 2 inhibitor, on duodenal polyposis in familial adenomatous polyposis. Gut 50 (6): 857-60, 2002. [PUBMED Abstract]
- Nugent KP, Farmer KC, Spigelman AD, et al.: Randomized controlled trial of the effect of sulindac on duodenal and rectal polyposis and cell proliferation in patients with familial adenomatous polyposis. Br J Surg 80 (12): 1618-9, 1993. [PUBMED Abstract]
- Jacoby RF, Cole CE, Hawk ET, et al.: Ursodeoxycholate/Sulindac combination treatment effectively prevents intestinal adenomas in a mouse model of polyposis. Gastroenterology 127 (3): 838-44, 2004. [PUBMED Abstract]
- Parc Y, Desaint B, Fléjou JF, et al.: The effect of ursodesoxycholic acid on duodenal adenomas in familial adenomatous polyposis: a prospective randomized placebo-control trial. Colorectal Dis 14 (7): 854-60, 2012. [PUBMED Abstract]
- van Heumen BW, Roelofs HM, Vink-Börger ME, et al.: Ursodeoxycholic acid counteracts celecoxib in reduction of duodenal polyps in patients with familial adenomatous polyposis: a multicentre, randomized controlled trial. Orphanet J Rare Dis 8: 118, 2013. [PUBMED Abstract]
- Fitzgerald GA: Coxibs and cardiovascular disease. N Engl J Med 351 (17): 1709-11, 2004. [PUBMED Abstract]
- Solomon SD, McMurray JJ, Pfeffer MA, et al.: Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N Engl J Med 352 (11): 1071-80, 2005. [PUBMED Abstract]
- Bresalier RS, Sandler RS, Quan H, et al.: Cardiovascular events associated with rofecoxib in a colorectal adenoma chemoprevention trial. N Engl J Med 352 (11): 1092-102, 2005. [PUBMED Abstract]
- Giardiello FM, Hamilton SR, Krush AJ, et al.: Treatment of colonic and rectal adenomas with sulindac in familial adenomatous polyposis. N Engl J Med 328 (18): 1313-6, 1993. [PUBMED Abstract]
- Roberts RB, Min L, Washington MK, et al.: Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc Natl Acad Sci U S A 99 (3): 1521-6, 2002. [PUBMED Abstract]
- Samadder NJ, Neklason DW, Boucher KM, et al.: Effect of Sulindac and Erlotinib vs Placebo on Duodenal Neoplasia in Familial Adenomatous Polyposis: A Randomized Clinical Trial. JAMA 315 (12): 1266-75, 2016 Mar 22-29. [PUBMED Abstract]
- Samadder NJ, Kuwada SK, Boucher KM, et al.: Association of Sulindac and Erlotinib vs Placebo With Colorectal Neoplasia in Familial Adenomatous Polyposis: Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol 4 (5): 671-677, 2018. [PUBMED Abstract]
- Rinella ES, Threadgill DW: Efficacy of EGFR inhibition is modulated by model, sex, genetic background and diet: implications for preclinical cancer prevention and therapy trials. PLoS One 7 (6): e39552, 2012. [PUBMED Abstract]
- Tonelli F, Ficari F, Valanzano R, et al.: Treatment of desmoids and mesenteric fibromatosis in familial adenomatous polyposis with raloxifene. Tumori 89 (4): 391-6, 2003 Jul-Aug. [PUBMED Abstract]
- Hansmann A, Adolph C, Vogel T, et al.: High-dose tamoxifen and sulindac as first-line treatment for desmoid tumors. Cancer 100 (3): 612-20, 2004. [PUBMED Abstract]
- Lindor NM, Dozois R, Nelson H, et al.: Desmoid tumors in familial adenomatous polyposis: a pilot project evaluating efficacy of treatment with pirfenidone. Am J Gastroenterol 98 (8): 1868-74, 2003. [PUBMED Abstract]
- Mace J, Sybil Biermann J, Sondak V, et al.: Response of extraabdominal desmoid tumors to therapy with imatinib mesylate. Cancer 95 (11): 2373-9, 2002. [PUBMED Abstract]
- Penel N, Le Cesne A, Bui BN, et al.: Imatinib for progressive and recurrent aggressive fibromatosis (desmoid tumors): an FNCLCC/French Sarcoma Group phase II trial with a long-term follow-up. Ann Oncol 22 (2): 452-7, 2011. [PUBMED Abstract]
- Kasper B, Gruenwald V, Reichardt P, et al.: Imatinib induces sustained progression arrest in RECIST progressive desmoid tumours: Final results of a phase II study of the German Interdisciplinary Sarcoma Group (GISG). Eur J Cancer 76: 60-67, 2017. [PUBMED Abstract]
- Gounder MM, Mahoney MR, Van Tine BA, et al.: Sorafenib for Advanced and Refractory Desmoid Tumors. N Engl J Med 379 (25): 2417-2428, 2018. [PUBMED Abstract]
- Latchford AR, Sturt NJ, Neale K, et al.: A 10-year review of surgery for desmoid disease associated with familial adenomatous polyposis. Br J Surg 93 (10): 1258-64, 2006. [PUBMED Abstract]
- Heiskanen I, Järvinen HJ: Occurrence of desmoid tumours in familial adenomatous polyposis and results of treatment. Int J Colorectal Dis 11 (4): 157-62, 1996. [PUBMED Abstract]
- Wood LD, Salaria SN, Cruise MW, et al.: Upper GI tract lesions in familial adenomatous polyposis (FAP): enrichment of pyloric gland adenomas and other gastric and duodenal neoplasms. Am J Surg Pathol 38 (3): 389-93, 2014. [PUBMED Abstract]
- Nakamura K, Nonaka S, Nakajima T, et al.: Clinical outcomes of gastric polyps and neoplasms in patients with familial adenomatous polyposis. Endosc Int Open 5 (3): E137-E145, 2017. [PUBMED Abstract]
- Park JS, Choi GS, Kim HJ, et al.: Natural orifice specimen extraction versus conventional laparoscopically assisted right hemicolectomy. Br J Surg 98 (5): 710-5, 2011. [PUBMED Abstract]
- Johnson MD, Mackey R, Brown N, et al.: Outcome based on management for duodenal adenomas: sporadic versus familial disease. J Gastrointest Surg 14 (2): 229-35, 2010. [PUBMED Abstract]
- de Vos tot Nederveen Cappel WH, Järvinen HJ, Björk J, et al.: Worldwide survey among polyposis registries of surgical management of severe duodenal adenomatosis in familial adenomatous polyposis. Br J Surg 90 (6): 705-10, 2003. [PUBMED Abstract]
- Balmaña J, Balaguer F, Cervantes A, et al.: Familial risk-colorectal cancer: ESMO Clinical Practice Guidelines. Ann Oncol 24 (Suppl 6): vi73-80, 2013. [PUBMED Abstract]
- Bülow S, Christensen IJ, Højen H, et al.: Duodenal surveillance improves the prognosis after duodenal cancer in familial adenomatous polyposis. Colorectal Dis 14 (8): 947-52, 2012. [PUBMED Abstract]
- Ahmad NA, Kochman ML, Long WB, et al.: Efficacy, safety, and clinical outcomes of endoscopic mucosal resection: a study of 101 cases. Gastrointest Endosc 55 (3): 390-6, 2002. [PUBMED Abstract]
- Heiskanen I, Kellokumpu I, Järvinen H: Management of duodenal adenomas in 98 patients with familial adenomatous polyposis. Endoscopy 31 (6): 412-6, 1999. [PUBMED Abstract]
- Penna C, Phillips RK, Tiret E, et al.: Surgical polypectomy of duodenal adenomas in familial adenomatous polyposis: experience of two European centres. Br J Surg 80 (8): 1027-9, 1993. [PUBMED Abstract]
- Mackey R, Walsh RM, Chung R, et al.: Pancreas-sparing duodenectomy is effective management for familial adenomatous polyposis. J Gastrointest Surg 9 (8): 1088-93; discussion 1093, 2005. [PUBMED Abstract]
- Collard MK, Lefevre JH, Ahmed O, et al.: Ten-year impact of pancreaticoduodenectomy on bowel function and quality of life of patients with ileal pouch-anal anastomosis for familial adenomatous polyposis. HPB (Oxford) 22 (10): 1402-1410, 2020. [PUBMED Abstract]
- Lepistö A, Kiviluoto T, Halttunen J, et al.: Surveillance and treatment of duodenal adenomatosis in familial adenomatous polyposis. Endoscopy 41 (6): 504-9, 2009. [PUBMED Abstract]
- Wallace MH, Phillips RK: Upper gastrointestinal disease in patients with familial adenomatous polyposis. Br J Surg 85 (6): 742-50, 1998. [PUBMED Abstract]
- Parc Y, Mabrut JY, Shields C, et al.: Surgical management of the duodenal manifestations of familial adenomatous polyposis. Br J Surg 98 (4): 480-4, 2011. [PUBMED Abstract]
- Penna C, Bataille N, Balladur P, et al.: Surgical treatment of severe duodenal polyposis in familial adenomatous polyposis. Br J Surg 85 (5): 665-8, 1998. [PUBMED Abstract]
- Hirasawa R, Iishi H, Tatsuta M, et al.: Clinicopathologic features and endoscopic resection of duodenal adenocarcinomas and adenomas with the submucosal saline injection technique. Gastrointest Endosc 46 (6): 507-13, 1997. [PUBMED Abstract]
- Catalano MF, Linder JD, Chak A, et al.: Endoscopic management of adenoma of the major duodenal papilla. Gastrointest Endosc 59 (2): 225-32, 2004. [PUBMED Abstract]
- Alarcon FJ, Burke CA, Church JM, et al.: Familial adenomatous polyposis: efficacy of endoscopic and surgical treatment for advanced duodenal adenomas. Dis Colon Rectum 42 (12): 1533-6, 1999. [PUBMED Abstract]
- Biasco G, Nobili E, Calabrese C, et al.: Impact of surgery on the development of duodenal cancer in patients with familial adenomatous polyposis. Dis Colon Rectum 49 (12): 1860-6, 2006. [PUBMED Abstract]
- Chung RS, Church JM, vanStolk R: Pancreas-sparing duodenectomy: indications, surgical technique, and results. Surgery 117 (3): 254-9, 1995. [PUBMED Abstract]
- Tsiotos GG, Sarr MG: Pancreas-preserving total duodenectomy. Dig Surg 15 (5): 398-403, 1998. [PUBMED Abstract]
- Sarmiento JM, Thompson GB, Nagorney DM, et al.: Pancreas-sparing duodenectomy for duodenal polyposis. Arch Surg 137 (5): 557-62; discussion 562-3, 2002. [PUBMED Abstract]
- Kalady MF, Clary BM, Tyler DS, et al.: Pancreas-preserving duodenectomy in the management of duodenal familial adenomatous polyposis. J Gastrointest Surg 6 (1): 82-7, 2002 Jan-Feb. [PUBMED Abstract]
- Eisenberger CF, Knoefel WT, Peiper M, et al.: Pancreas-sparing duodenectomy in duodenal pathology: indications and results. Hepatogastroenterology 51 (57): 727-31, 2004 May-Jun. [PUBMED Abstract]
- Jasperson KW, Tuohy TM, Neklason DW, et al.: Hereditary and familial colon cancer. Gastroenterology 138 (6): 2044-58, 2010. [PUBMED Abstract]
- Jarrar AM, Milas M, Mitchell J, et al.: Screening for thyroid cancer in patients with familial adenomatous polyposis. Ann Surg 253 (3): 515-21, 2011. [PUBMED Abstract]
- Aretz S, Koch A, Uhlhaas S, et al.: Should children at risk for familial adenomatous polyposis be screened for hepatoblastoma and children with apparently sporadic hepatoblastoma be screened for APC germline mutations? Pediatr Blood Cancer 47 (6): 811-8, 2006. [PUBMED Abstract]
- Leppert M, Burt R, Hughes JP, et al.: Genetic analysis of an inherited predisposition to colon cancer in a family with a variable number of adenomatous polyps. N Engl J Med 322 (13): 904-8, 1990. [PUBMED Abstract]
- Lynch HT, Smyrk TC: Classification of familial adenomatous polyposis: a diagnostic nightmare. Am J Hum Genet 62 (6): 1288-9, 1998. [PUBMED Abstract]
- Giardiello FM, Brensinger JD, Luce MC, et al.: Phenotypic expression of disease in families that have mutations in the 5' region of the adenomatous polyposis coli gene. Ann Intern Med 126 (7): 514-9, 1997. [PUBMED Abstract]
- White S, Bubb VJ, Wyllie AH: Germline APC mutation (Gln1317) in a cancer-prone family that does not result in familial adenomatous polyposis. Genes Chromosomes Cancer 15 (2): 122-8, 1996. [PUBMED Abstract]
- Gonçalves V, Theisen P, Antunes O, et al.: A missense mutation in the APC tumor suppressor gene disrupts an ASF/SF2 splicing enhancer motif and causes pathogenic skipping of exon 14. Mutat Res 662 (1-2): 33-6, 2009. [PUBMED Abstract]
- Heppner Goss K, Trzepacz C, Tuohy TM, et al.: Attenuated APC alleles produce functional protein from internal translation initiation. Proc Natl Acad Sci U S A 99 (12): 8161-6, 2002. [PUBMED Abstract]
- Nieuwenhuis MH, Vasen HF: Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol 61 (2): 153-61, 2007. [PUBMED Abstract]
- Scott RJ, Meldrum C, Crooks R, et al.: Familial adenomatous polyposis: more evidence for disease diversity and genetic heterogeneity. Gut 48 (4): 508-14, 2001. [PUBMED Abstract]
- Knudsen AL, Bisgaard ML, Bülow S: Attenuated familial adenomatous polyposis (AFAP). A review of the literature. Fam Cancer 2 (1): 43-55, 2003. [PUBMED Abstract]
- Vasen HF, Möslein G, Alonso A, et al.: Guidelines for the clinical management of familial adenomatous polyposis (FAP). Gut 57 (5): 704-13, 2008. [PUBMED Abstract]
- Nielsen M, Morreau H, Vasen HF, et al.: MUTYH-associated polyposis (MAP). Crit Rev Oncol Hematol 79 (1): 1-16, 2011. [PUBMED Abstract]
- Nielsen M, Joerink-van de Beld MC, Jones N, et al.: Analysis of MUTYH genotypes and colorectal phenotypes in patients With MUTYH-associated polyposis. Gastroenterology 136 (2): 471-6, 2009. [PUBMED Abstract]
- Nielsen M, Franken PF, Reinards TH, et al.: Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP). J Med Genet 42 (9): e54, 2005. [PUBMED Abstract]
- Knopperts AP, Nielsen M, Niessen RC, et al.: Contribution of bi-allelic germline MUTYH mutations to early-onset and familial colorectal cancer and to low number of adenomatous polyps: case-series and literature review. Fam Cancer 12 (1): 43-50, 2013. [PUBMED Abstract]
- Sampson JR, Dolwani S, Jones S, et al.: Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet 362 (9377): 39-41, 2003. [PUBMED Abstract]
- Dolwani S, Williams GT, West KP, et al.: Analysis of inherited MYH/(MutYH) mutations in British Asian patients with colorectal cancer. Gut 56 (4): 593, 2007. [PUBMED Abstract]
- Gismondi V, Meta M, Bonelli L, et al.: Prevalence of the Y165C, G382D and 1395delGGA germline mutations of the MYH gene in Italian patients with adenomatous polyposis coli and colorectal adenomas. Int J Cancer 109 (5): 680-4, 2004. [PUBMED Abstract]
- Ricci MT, Miccoli S, Turchetti D, et al.: Type and frequency of MUTYH variants in Italian patients with suspected MAP: a retrospective multicenter study. J Hum Genet 62 (2): 309-315, 2017. [PUBMED Abstract]
- Isidro G, Laranjeira F, Pires A, et al.: Germline MUTYH (MYH) mutations in Portuguese individuals with multiple colorectal adenomas. Hum Mutat 24 (4): 353-4, 2004. [PUBMED Abstract]
- Kim DW, Kim IJ, Kang HC, et al.: Germline mutations of the MYH gene in Korean patients with multiple colorectal adenomas. Int J Colorectal Dis 22 (10): 1173-8, 2007. [PUBMED Abstract]
- Yanaru-Fujisawa R, Matsumoto T, Ushijima Y, et al.: Genomic and functional analyses of MUTYH in Japanese patients with adenomatous polyposis. Clin Genet 73 (6): 545-53, 2008. [PUBMED Abstract]
- Kim JC, Ka IH, Lee YM, et al.: MYH, OGG1, MTH1, and APC alterations involved in the colorectal tumorigenesis of Korean patients with multiple adenomas. Virchows Arch 450 (3): 311-9, 2007. [PUBMED Abstract]
- Hampel H: Genetic testing for hereditary colorectal cancer. Surg Oncol Clin N Am 18 (4): 687-703, 2009. [PUBMED Abstract]
- Jones N, Vogt S, Nielsen M, et al.: Increased colorectal cancer incidence in obligate carriers of heterozygous mutations in MUTYH. Gastroenterology 137 (2): 489-94, 494.e1; quiz 725-6, 2009. [PUBMED Abstract]
- Sieber OM, Lipton L, Crabtree M, et al.: Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med 348 (9): 791-9, 2003. [PUBMED Abstract]
- Nieuwenhuis MH, Vogt S, Jones N, et al.: Evidence for accelerated colorectal adenoma--carcinoma progression in MUTYH-associated polyposis? Gut 61 (5): 734-8, 2012. [PUBMED Abstract]
- Win AK, Dowty JG, Cleary SP, et al.: Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology 146 (5): 1208-11.e1-5, 2014. [PUBMED Abstract]
- Morak M, Laner A, Bacher U, et al.: MUTYH-associated polyposis - variability of the clinical phenotype in patients with biallelic and monoallelic MUTYH mutations and report on novel mutations. Clin Genet 78 (4): 353-63, 2010. [PUBMED Abstract]
- Boparai KS, Dekker E, Van Eeden S, et al.: Hyperplastic polyps and sessile serrated adenomas as a phenotypic expression of MYH-associated polyposis. Gastroenterology 135 (6): 2014-8, 2008. [PUBMED Abstract]
- Nascimbeni R, Pucciarelli S, Di Lorenzo D, et al.: Rectum-sparing surgery may be appropriate for biallelic MutYH-associated polyposis. Dis Colon Rectum 53 (12): 1670-5, 2010. [PUBMED Abstract]
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Colorectal Cancer Screening. Version 1.2024. Plymouth Meeting, PA: National Comprehensive Cancer Network, 2024. Available with free registration. Last accessed March 3, 2024.
- Win AK, Cleary SP, Dowty JG, et al.: Cancer risks for monoallelic MUTYH mutation carriers with a family history of colorectal cancer. Int J Cancer 129 (9): 2256-62, 2011. [PUBMED Abstract]
- Vogt S, Jones N, Christian D, et al.: Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology 137 (6): 1976-85.e1-10, 2009. [PUBMED Abstract]
- Lefevre JH, Rodrigue CM, Mourra N, et al.: Implication of MYH in colorectal polyposis. Ann Surg 244 (6): 874-9; discussion 879-80, 2006. [PUBMED Abstract]
- Wasielewski M, Out AA, Vermeulen J, et al.: Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat 124 (3): 635-41, 2010. [PUBMED Abstract]
- Poulsen ML, Bisgaard ML: MUTYH Associated Polyposis (MAP). Curr Genomics 9 (6): 420-35, 2008. [PUBMED Abstract]
- Goodenberger M, Lindor NM: Lynch syndrome and MYH-associated polyposis: review and testing strategy. J Clin Gastroenterol 45 (6): 488-500, 2011. [PUBMED Abstract]
- Walton SJ, Kallenberg FG, Clark SK, et al.: Frequency and Features of Duodenal Adenomas in Patients With MUTYH-Associated Polyposis. Clin Gastroenterol Hepatol 14 (7): 986-92, 2016. [PUBMED Abstract]
- Win AK, Hopper JL, Jenkins MA: Association between monoallelic MUTYH mutation and colorectal cancer risk: a meta-regression analysis. Fam Cancer 10 (1): 1-9, 2011. [PUBMED Abstract]
- Giráldez MD, Balaguer F, Caldés T, et al.: Association of MUTYH and MSH6 germline mutations in colorectal cancer patients. Fam Cancer 8 (4): 525-31, 2009. [PUBMED Abstract]
- Steinke V, Rahner N, Morak M, et al.: No association between MUTYH and MSH6 germline mutations in 64 HNPCC patients. Eur J Hum Genet 16 (5): 587-92, 2008. [PUBMED Abstract]
- Win AK, Reece JC, Buchanan DD, et al.: Risk of colorectal cancer for people with a mutation in both a MUTYH and a DNA mismatch repair gene. Fam Cancer 14 (4): 575-83, 2015. [PUBMED Abstract]
- Boland CR, Troncale FJ: Familial colonic cancer without antecedent polyposis. Ann Intern Med 100 (5): 700-1, 1984. [PUBMED Abstract]
- Vasen HF, Mecklin JP, Khan PM, et al.: The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 34 (5): 424-5, 1991. [PUBMED Abstract]
- Bodmer WF, Bailey CJ, Bodmer J, et al.: Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature 328 (6131): 614-6, 1987 Aug 13-19. [PUBMED Abstract]
- Groden J, Thliveris A, Samowitz W, et al.: Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66 (3): 589-600, 1991. [PUBMED Abstract]
- Vasen HF, Watson P, Mecklin JP, et al.: New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116 (6): 1453-6, 1999. [PUBMED Abstract]
- Lindor NM, Rabe K, Petersen GM, et al.: Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA 293 (16): 1979-85, 2005. [PUBMED Abstract]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, et al.: A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89 (23): 1758-62, 1997. [PUBMED Abstract]
- Umar A, Boland CR, Terdiman JP, et al.: Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 96 (4): 261-8, 2004. [PUBMED Abstract]
- Laghi L, Bianchi P, Roncalli M, et al.: Re: Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 96 (18): 1402-3; author reply 1403-4, 2004. [PUBMED Abstract]
- Hampel H, Frankel WL, Martin E, et al.: Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 26 (35): 5783-8, 2008. [PUBMED Abstract]
- Grover S, Stoffel EM, Bussone L, et al.: Physician assessment of family cancer history and referral for genetic evaluation in colorectal cancer patients. Clin Gastroenterol Hepatol 2 (9): 813-9, 2004. [PUBMED Abstract]
- Barnetson RA, Tenesa A, Farrington SM, et al.: Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med 354 (26): 2751-63, 2006. [PUBMED Abstract]
- Kastrinos F, Steyerberg EW, Mercado R, et al.: The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology 140 (1): 73-81, 2011. [PUBMED Abstract]
- Chen S, Wang W, Lee S, et al.: Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 296 (12): 1479-87, 2006. [PUBMED Abstract]
- Kastrinos F, Uno H, Ukaegbu C, et al.: Development and Validation of the PREMM5 Model for Comprehensive Risk Assessment of Lynch Syndrome. J Clin Oncol 35 (19): 2165-2172, 2017. [PUBMED Abstract]
- Yurgelun MB, Uno H, Furniss CS, et al.: Development and Validation of the PREMMplus Model for Multigene Hereditary Cancer Risk Assessment. J Clin Oncol 40 (35): 4083-4094, 2022. [PUBMED Abstract]
- Kastrinos F, Allen JI, Stockwell DH, et al.: Development and validation of a colon cancer risk assessment tool for patients undergoing colonoscopy. Am J Gastroenterol 104 (6): 1508-18, 2009. [PUBMED Abstract]
- Balaguer F, Balmaña J, Castellví-Bel S, et al.: Validation and extension of the PREMM1,2 model in a population-based cohort of colorectal cancer patients. Gastroenterology 134 (1): 39-46, 2008. [PUBMED Abstract]
- Balmaña J, Balaguer F, Castellví-Bel S, et al.: Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet 45 (9): 557-63, 2008. [PUBMED Abstract]
- Green RC, Parfrey PS, Woods MO, et al.: Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst 101 (5): 331-40, 2009. [PUBMED Abstract]
- Kastrinos F, Steyerberg EW, Balmaña J, et al.: Comparison of the clinical prediction model PREMM(1,2,6) and molecular testing for the systematic identification of Lynch syndrome in colorectal cancer. Gut 62 (2): 272-9, 2013. [PUBMED Abstract]
- Khan O, Blanco A, Conrad P, et al.: Performance of Lynch syndrome predictive models in a multi-center US referral population. Am J Gastroenterol 106 (10): 1822-7; quiz 1828, 2011. [PUBMED Abstract]
- Pouchet CJ, Wong N, Chong G, et al.: A comparison of models used to predict MLH1, MSH2 and MSH6 mutation carriers. Ann Oncol 20 (4): 681-8, 2009. [PUBMED Abstract]
- Monzon JG, Cremin C, Armstrong L, et al.: Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer 126 (4): 930-9, 2010. [PUBMED Abstract]
- Kastrinos F, Balmaña J, Syngal S: Prediction models in Lynch syndrome. Fam Cancer 12 (2): 217-28, 2013. [PUBMED Abstract]
- Balmaña J, Stockwell DH, Steyerberg EW, et al.: Prediction of MLH1 and MSH2 mutations in Lynch syndrome. JAMA 296 (12): 1469-78, 2006. [PUBMED Abstract]
- Luba DG, DiSario JA, Rock C, et al.: Community Practice Implementation of a Self-administered Version of PREMM1,2,6 to Assess Risk for Lynch Syndrome. Clin Gastroenterol Hepatol 16 (1): 49-58, 2018. [PUBMED Abstract]
- Kastrinos F, Ojha RP, Leenen C, et al.: Comparison of Prediction Models for Lynch Syndrome Among Individuals With Colorectal Cancer. J Natl Cancer Inst 108 (2): , 2016. [PUBMED Abstract]
- Weber JL, May PE: Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44 (3): 388-96, 1989. [PUBMED Abstract]
- Vilar E, Gruber SB: Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 7 (3): 153-62, 2010. [PUBMED Abstract]
- Haraldsdottir S, Roth R, Pearlman R, et al.: Mismatch repair deficiency concordance between primary colorectal cancer and corresponding metastasis. Fam Cancer 15 (2): 253-60, 2016. [PUBMED Abstract]
- Grady WM, Carethers JM: Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135 (4): 1079-99, 2008. [PUBMED Abstract]
- Greenson JK, Huang SC, Herron C, et al.: Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol 33 (1): 126-33, 2009. [PUBMED Abstract]
- Boland CR, Thibodeau SN, Hamilton SR, et al.: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58 (22): 5248-57, 1998. [PUBMED Abstract]
- Thibodeau SN, French AJ, Roche PC, et al.: Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 56 (21): 4836-40, 1996. [PUBMED Abstract]
- Cawkwell L, Gray S, Murgatroyd H, et al.: Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut 45 (3): 409-15, 1999. [PUBMED Abstract]
- Lindor NM, Burgart LJ, Leontovich O, et al.: Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 20 (4): 1043-8, 2002. [PUBMED Abstract]
- de La Chapelle A: Microsatellite instability phenotype of tumors: genotyping or immunohistochemistry? The jury is still out. J Clin Oncol 20 (4): 897-9, 2002. [PUBMED Abstract]
- Peltomäki P: Update on Lynch syndrome genomics. Fam Cancer 15 (3): 385-93, 2016. [PUBMED Abstract]
- Rumilla K, Schowalter KV, Lindor NM, et al.: Frequency of deletions of EPCAM (TACSTD1) in MSH2-associated Lynch syndrome cases. J Mol Diagn 13 (1): 93-9, 2011. [PUBMED Abstract]
- Kovacs ME, Papp J, Szentirmay Z, et al.: Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat 30 (2): 197-203, 2009. [PUBMED Abstract]
- Rosty C, Clendenning M, Walsh MD, et al.: Germline mutations in PMS2 and MLH1 in individuals with solitary loss of PMS2 expression in colorectal carcinomas from the Colon Cancer Family Registry Cohort. BMJ Open 6 (2): e010293, 2016. [PUBMED Abstract]
- Lynch HT, Boland CR, Rodriguez-Bigas MA, et al.: Who should be sent for genetic testing in hereditary colorectal cancer syndromes? J Clin Oncol 25 (23): 3534-42, 2007. [PUBMED Abstract]
- Cunningham JM, Kim CY, Christensen ER, et al.: The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet 69 (4): 780-90, 2001. [PUBMED Abstract]
- Esteller M: Epigenetics in cancer. N Engl J Med 358 (11): 1148-59, 2008. [PUBMED Abstract]
- Wang L, Cunningham JM, Winters JL, et al.: BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 63 (17): 5209-12, 2003. [PUBMED Abstract]
- Domingo E, Espín E, Armengol M, et al.: Activated BRAF targets proximal colon tumors with mismatch repair deficiency and MLH1 inactivation. Genes Chromosomes Cancer 39 (2): 138-42, 2004. [PUBMED Abstract]
- Deng G, Bell I, Crawley S, et al.: BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res 10 (1 Pt 1): 191-5, 2004. [PUBMED Abstract]
- Domingo E, Niessen RC, Oliveira C, et al.: BRAF-V600E is not involved in the colorectal tumorigenesis of HNPCC in patients with functional MLH1 and MSH2 genes. Oncogene 24 (24): 3995-8, 2005. [PUBMED Abstract]
- Haraldsdottir S, Hampel H, Tomsic J, et al.: Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology 147 (6): 1308-1316.e1, 2014. [PUBMED Abstract]
- Pearlman R, Frankel WL, Swanson BJ, et al.: Prospective Statewide Study of Universal Screening for Hereditary Colorectal Cancer: The Ohio Colorectal Cancer Prevention Initiative. JCO Precis Oncol 5: , 2021. [PUBMED Abstract]
- Hampel H, Pearlman R, de la Chapelle A, et al.: Double somatic mismatch repair gene pathogenic variants as common as Lynch syndrome among endometrial cancer patients. Gynecol Oncol 160 (1): 161-168, 2021. [PUBMED Abstract]
- Dixon K, Asrat MJ, Bedard AC, et al.: Integrating Tumor Sequencing Into Clinical Practice for Patients With Mismatch Repair-Deficient Lynch Syndrome Spectrum Cancers. Clin Transl Gastroenterol 12 (8): e00397, 2021. [PUBMED Abstract]
- Antelo M, Golubicki M, Roca E, et al.: Lynch-like syndrome is as frequent as Lynch syndrome in early-onset nonfamilial nonpolyposis colorectal cancer. Int J Cancer 145 (3): 705-713, 2019. [PUBMED Abstract]
- Carethers JM, Stoffel EM: Lynch syndrome and Lynch syndrome mimics: The growing complex landscape of hereditary colon cancer. World J Gastroenterol 21 (31): 9253-61, 2015. [PUBMED Abstract]
- Bittles AH, Black ML: Evolution in health and medicine Sackler colloquium: Consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci U S A 107 (Suppl 1): 1779-86, 2010. [PUBMED Abstract]
- Hitchins MP: The role of epigenetics in Lynch syndrome. Fam Cancer 12 (2): 189-205, 2013. [PUBMED Abstract]
- Gazzoli I, Loda M, Garber J, et al.: A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res 62 (14): 3925-8, 2002. [PUBMED Abstract]
- Gylling A, Ridanpää M, Vierimaa O, et al.: Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer 124 (10): 2333-40, 2009. [PUBMED Abstract]
- Hitchins MP, Rapkins RW, Kwok CT, et al.: Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5'UTR. Cancer Cell 20 (2): 200-13, 2011. [PUBMED Abstract]
- Goel A, Nguyen TP, Leung HC, et al.: De novo constitutional MLH1 epimutations confer early-onset colorectal cancer in two new sporadic Lynch syndrome cases, with derivation of the epimutation on the paternal allele in one. Int J Cancer 128 (4): 869-78, 2011. [PUBMED Abstract]
- Hitchins MP, Wong JJ, Suthers G, et al.: Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med 356 (7): 697-705, 2007. [PUBMED Abstract]
- Hampel H, Frankel WL, Martin E, et al.: Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352 (18): 1851-60, 2005. [PUBMED Abstract]
- Ladabaum U, Wang G, Terdiman J, et al.: Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med 155 (2): 69-79, 2011. [PUBMED Abstract]
- Piñol V, Castells A, Andreu M, et al.: Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA 293 (16): 1986-94, 2005. [PUBMED Abstract]
- Baudhuin LM, Burgart LJ, Leontovich O, et al.: Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer 4 (3): 255-65, 2005. [PUBMED Abstract]
- Lagerstedt Robinson K, Liu T, Vandrovcova J, et al.: Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst 99 (4): 291-9, 2007. [PUBMED Abstract]
- Schofield L, Watson N, Grieu F, et al.: Population-based detection of Lynch syndrome in young colorectal cancer patients using microsatellite instability as the initial test. Int J Cancer 124 (5): 1097-102, 2009. [PUBMED Abstract]
- Mills AM, Liou S, Ford JM, et al.: Lynch syndrome screening should be considered for all patients with newly diagnosed endometrial cancer. Am J Surg Pathol 38 (11): 1501-9, 2014. [PUBMED Abstract]
- Giardiello FM, Allen JI, Axilbund JE, et al.: Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol 109 (8): 1159-79, 2014. [PUBMED Abstract]
- Rubenstein JH, Enns R, Heidelbaugh J, et al.: American Gastroenterological Association Institute Guideline on the Diagnosis and Management of Lynch Syndrome. Gastroenterology 149 (3): 777-82; quiz e16-7, 2015. [PUBMED Abstract]
- Committee on Practice Bulletins-Gynecology, Society of Gynecologic Oncology: ACOG Practice Bulletin No. 147: Lynch syndrome. Obstet Gynecol 124 (5): 1042-54, 2014. [PUBMED Abstract]
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group: Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 11 (1): 35-41, 2009. [PUBMED Abstract]
- Palomaki GE, McClain MR, Melillo S, et al.: EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med 11 (1): 42-65, 2009. [PUBMED Abstract]
- Moreira L, Balaguer F, Lindor N, et al.: Identification of Lynch syndrome among patients with colorectal cancer. JAMA 308 (15): 1555-65, 2012. [PUBMED Abstract]
- Boland CR, Shike M: Report from the Jerusalem workshop on Lynch syndrome-hereditary nonpolyposis colorectal cancer. Gastroenterology 138 (7): 2197.e1-7, 2010. [PUBMED Abstract]
- Crucianelli F, Tricarico R, Turchetti D, et al.: MLH1 constitutional and somatic methylation in patients with MLH1 negative tumors fulfilling the revised Bethesda criteria. Epigenetics 9 (10): 1431-8, 2014. [PUBMED Abstract]
- Leenen CH, Goverde A, de Bekker-Grob EW, et al.: Cost-effectiveness of routine screening for Lynch syndrome in colorectal cancer patients up to 70 years of age. Genet Med 18 (10): 966-73, 2016. [PUBMED Abstract]
- Hampel H, Pearlman R, Beightol M, et al.: Assessment of Tumor Sequencing as a Replacement for Lynch Syndrome Screening and Current Molecular Tests for Patients With Colorectal Cancer. JAMA Oncol 4 (6): 806-813, 2018. [PUBMED Abstract]
- Li D, Hoodfar E, Jiang SF, et al.: Comparison of Universal Versus Age-Restricted Screening of Colorectal Tumors for Lynch Syndrome Using Mismatch Repair Immunohistochemistry: A Cohort Study. Ann Intern Med 171 (1): 19-26, 2019. [PUBMED Abstract]
- Barzi A, Sadeghi S, Kattan MW, et al.: Comparative effectiveness of screening strategies for Lynch syndrome. J Natl Cancer Inst 107 (4): , 2015. [PUBMED Abstract]
- Stoffel EM, Mangu PB, Gruber SB, et al.: Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J Clin Oncol 33 (2): 209-17, 2015. [PUBMED Abstract]
- Goverde A, Spaander MC, van Doorn HC, et al.: Cost-effectiveness of routine screening for Lynch syndrome in endometrial cancer patients up to 70years of age. Gynecol Oncol 143 (3): 453-459, 2016. [PUBMED Abstract]
- Cohen SA: Current Lynch syndrome tumor screening practices: a survey of genetic counselors. J Genet Couns 23 (1): 38-47, 2014. [PUBMED Abstract]
- Beamer LC, Grant ML, Espenschied CR, et al.: Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol 30 (10): 1058-63, 2012. [PUBMED Abstract]
- Dineen S, Lynch PM, Rodriguez-Bigas MA, et al.: A Prospective Six Sigma Quality Improvement Trial to Optimize Universal Screening for Genetic Syndrome Among Patients With Young-Onset Colorectal Cancer. J Natl Compr Canc Netw 13 (7): 865-72, 2015. [PUBMED Abstract]
- Heald B, Plesec T, Liu X, et al.: Implementation of universal microsatellite instability and immunohistochemistry screening for diagnosing lynch syndrome in a large academic medical center. J Clin Oncol 31 (10): 1336-40, 2013. [PUBMED Abstract]
- Cragun D, DeBate RD, Vadaparampil ST, et al.: Comparing universal Lynch syndrome tumor-screening programs to evaluate associations between implementation strategies and patient follow-through. Genet Med 16 (10): 773-82, 2014. [PUBMED Abstract]
- Ward RL, Hicks S, Hawkins NJ: Population-based molecular screening for Lynch syndrome: implications for personalized medicine. J Clin Oncol 31 (20): 2554-62, 2013. [PUBMED Abstract]
- Hampel H, Frankel W, Panescu J, et al.: Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res 66 (15): 7810-7, 2006. [PUBMED Abstract]
- Hampel H, Panescu J, Lockman J, et al.: Comment on: Screening for Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) among Endometrial Cancer Patients. Cancer Res 67 (19): 9603, 2007. [PUBMED Abstract]
- Watkins JC, Yang EJ, Muto MG, et al.: Universal Screening for Mismatch-Repair Deficiency in Endometrial Cancers to Identify Patients With Lynch Syndrome and Lynch-like Syndrome. Int J Gynecol Pathol 36 (2): 115-127, 2017. [PUBMED Abstract]
- Adar T, Rodgers LH, Shannon KM, et al.: Universal screening of both endometrial and colon cancers increases the detection of Lynch syndrome. Cancer 124 (15): 3145-3153, 2018. [PUBMED Abstract]
- Kwon JS, Scott JL, Gilks CB, et al.: Testing women with endometrial cancer to detect Lynch syndrome. J Clin Oncol 29 (16): 2247-52, 2011. [PUBMED Abstract]
- Latham A, Srinivasan P, Kemel Y, et al.: Microsatellite Instability Is Associated With the Presence of Lynch Syndrome Pan-Cancer. J Clin Oncol 37 (4): 286-295, 2019. [PUBMED Abstract]
- Middha S, Zhang L, Nafa K, et al.: Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precis Oncol 2017: , 2017. [PUBMED Abstract]
- Latham A, Shia J, Patel Z, et al.: Characterization and Clinical Outcomes of DNA Mismatch Repair-deficient Small Bowel Adenocarcinoma. Clin Cancer Res 27 (5): 1429-1437, 2021. [PUBMED Abstract]
- Pearlman R, Frankel WL, Swanson B, et al.: Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol 3 (4): 464-471, 2017. [PUBMED Abstract]
- Yurgelun MB, Allen B, Kaldate RR, et al.: Identification of a Variety of Mutations in Cancer Predisposition Genes in Patients With Suspected Lynch Syndrome. Gastroenterology 149 (3): 604-13.e20, 2015. [PUBMED Abstract]
- You YN, Borras E, Chang K, et al.: Detection of Pathogenic Germline Variants Among Patients With Advanced Colorectal Cancer Undergoing Tumor Genomic Profiling for Precision Medicine. Dis Colon Rectum 62 (4): 429-437, 2019. [PUBMED Abstract]
- Yurgelun MB, Kulke MH, Fuchs CS, et al.: Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer. J Clin Oncol 35 (10): 1086-1095, 2017. [PUBMED Abstract]
- Espenschied CR, LaDuca H, Li S, et al.: Multigene Panel Testing Provides a New Perspective on Lynch Syndrome. J Clin Oncol 35 (22): 2568-2575, 2017. [PUBMED Abstract]
- Roberts ME, Jackson SA, Susswein LR, et al.: MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet Med 20 (10): 1167-1174, 2018. [PUBMED Abstract]
- Neumann PJ, Cohen JT, Weinstein MC: Updating cost-effectiveness--the curious resilience of the $50,000-per-QALY threshold. N Engl J Med 371 (9): 796-7, 2014. [PUBMED Abstract]
- Gallego CJ, Shirts BH, Bennette CS, et al.: Next-Generation Sequencing Panels for the Diagnosis of Colorectal Cancer and Polyposis Syndromes: A Cost-Effectiveness Analysis. J Clin Oncol 33 (18): 2084-91, 2015. [PUBMED Abstract]
- Bronner CE, Baker SM, Morrison PT, et al.: Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368 (6468): 258-61, 1994. [PUBMED Abstract]
- Papadopoulos N, Nicolaides NC, Wei YF, et al.: Mutation of a mutL homolog in hereditary colon cancer. Science 263 (5153): 1625-9, 1994. [PUBMED Abstract]
- Fishel R, Lescoe MK, Rao MR, et al.: The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75 (5): 1027-38, 1993. [PUBMED Abstract]
- Leach FS, Nicolaides NC, Papadopoulos N, et al.: Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75 (6): 1215-25, 1993. [PUBMED Abstract]
- Miyaki M, Konishi M, Tanaka K, et al.: Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 17 (3): 271-2, 1997. [PUBMED Abstract]
- Nicolaides NC, Papadopoulos N, Liu B, et al.: Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371 (6492): 75-80, 1994. [PUBMED Abstract]
- Worthley DL, Walsh MD, Barker M, et al.: Familial mutations in PMS2 can cause autosomal dominant hereditary nonpolyposis colorectal cancer. Gastroenterology 128 (5): 1431-6, 2005. [PUBMED Abstract]
- Peltomäki P, Aaltonen LA, Sistonen P, et al.: Genetic mapping of a locus predisposing to human colorectal cancer. Science 260 (5109): 810-2, 1993. [PUBMED Abstract]
- Lindblom A, Tannergård P, Werelius B, et al.: Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat Genet 5 (3): 279-82, 1993. [PUBMED Abstract]
- Ligtenberg MJ, Kuiper RP, Chan TL, et al.: Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat Genet 41 (1): 112-7, 2009. [PUBMED Abstract]
- Kuiper RP, Vissers LE, Venkatachalam R, et al.: Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat 32 (4): 407-14, 2011. [PUBMED Abstract]
- Vasen HF: Clinical description of the Lynch syndrome [hereditary nonpolyposis colorectal cancer (HNPCC)]. Fam Cancer 4 (3): 219-25, 2005. [PUBMED Abstract]
- National Cancer Institute: SEER Stat Fact Sheets: Colorectal Cancer. Bethesda, Md: National Institutes of Health. Available online. Last accessed April 10, 2024.
- Hampel H, Stephens JA, Pukkala E, et al.: Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: later age of onset. Gastroenterology 129 (2): 415-21, 2005. [PUBMED Abstract]
- Win AK, Jenkins MA, Dowty JG, et al.: Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer. Cancer Epidemiol Biomarkers Prev 26 (3): 404-412, 2017. [PUBMED Abstract]
- Marra G, Boland CR: Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J Natl Cancer Inst 87 (15): 1114-25, 1995. [PUBMED Abstract]
- Peltomäki P, Vasen HF: Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113 (4): 1146-58, 1997. [PUBMED Abstract]
- Plazzer JP, Sijmons RH, Woods MO, et al.: The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam Cancer 12 (2): 175-80, 2013. [PUBMED Abstract]
- Peltomäki P: Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol 21 (6): 1174-9, 2003. [PUBMED Abstract]
- Vasen HF, Stormorken A, Menko FH, et al.: MSH2 mutation carriers are at higher risk of cancer than MLH1 mutation carriers: a study of hereditary nonpolyposis colorectal cancer families. J Clin Oncol 19 (20): 4074-80, 2001. [PUBMED Abstract]
- Ryan NAJ, Morris J, Green K, et al.: Association of Mismatch Repair Mutation With Age at Cancer Onset in Lynch Syndrome: Implications for Stratified Surveillance Strategies. JAMA Oncol 3 (12): 1702-1706, 2017. [PUBMED Abstract]
- Quehenberger F, Vasen HF, van Houwelingen HC: Risk of colorectal and endometrial cancer for carriers of mutations of the hMLH1 and hMSH2 gene: correction for ascertainment. J Med Genet 42 (6): 491-6, 2005. [PUBMED Abstract]
- Lin KM, Shashidharan M, Thorson AG, et al.: Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg 2 (1): 67-71, 1998 Jan-Feb. [PUBMED Abstract]
- Plaschke J, Engel C, Krüger S, et al.: Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: the German Hereditary Nonpolyposis Colorectal Cancer Consortium. J Clin Oncol 22 (22): 4486-94, 2004. [PUBMED Abstract]
- Berends MJ, Wu Y, Sijmons RH, et al.: Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet 70 (1): 26-37, 2002. [PUBMED Abstract]
- Ramsoekh D, Wagner A, van Leerdam ME, et al.: A high incidence of MSH6 mutations in Amsterdam criteria II-negative families tested in a diagnostic setting. Gut 57 (11): 1539-44, 2008. [PUBMED Abstract]
- Peltomäki P, Vasen H: Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers 20 (4-5): 269-76, 2004. [PUBMED Abstract]
- Kolodner RD, Tytell JD, Schmeits JL, et al.: Germ-line msh6 mutations in colorectal cancer families. Cancer Res 59 (20): 5068-74, 1999. [PUBMED Abstract]
- Peterlongo P, Nafa K, Lerman GS, et al.: MSH6 germline mutations are rare in colorectal cancer families. Int J Cancer 107 (4): 571-9, 2003. [PUBMED Abstract]
- Hendriks YM, Wagner A, Morreau H, et al.: Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology 127 (1): 17-25, 2004. [PUBMED Abstract]
- Goodenberger ML, Thomas BC, Riegert-Johnson D, et al.: PMS2 monoallelic mutation carriers: the known unknown. Genet Med 18 (1): 13-9, 2016. [PUBMED Abstract]
- Hendriks YM, Jagmohan-Changur S, van der Klift HM, et al.: Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome). Gastroenterology 130 (2): 312-22, 2006. [PUBMED Abstract]
- Truninger K, Menigatti M, Luz J, et al.: Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology 128 (5): 1160-71, 2005. [PUBMED Abstract]
- Senter L, Clendenning M, Sotamaa K, et al.: The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 135 (2): 419-28, 2008. [PUBMED Abstract]
- ten Broeke SW, Brohet RM, Tops CM, et al.: Lynch syndrome caused by germline PMS2 mutations: delineating the cancer risk. J Clin Oncol 33 (4): 319-25, 2015. [PUBMED Abstract]
- Ten Broeke SW, van der Klift HM, Tops CMJ, et al.: Cancer Risks for PMS2-Associated Lynch Syndrome. J Clin Oncol 36 (29): 2961-2968, 2018. [PUBMED Abstract]
- Møller P, Seppälä T, Bernstein I, et al.: Incidence of and survival after subsequent cancers in carriers of pathogenic MMR variants with previous cancer: a report from the prospective Lynch syndrome database. Gut 66 (9): 1657-1664, 2017. [PUBMED Abstract]
- Møller P, Seppälä TT, Bernstein I, et al.: Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 67 (7): 1306-1316, 2018. [PUBMED Abstract]
- Engel C, Vasen HF, Seppälä T, et al.: No Difference in Colorectal Cancer Incidence or Stage at Detection by Colonoscopy Among 3 Countries With Different Lynch Syndrome Surveillance Policies. Gastroenterology 155 (5): 1400-1409.e2, 2018. [PUBMED Abstract]
- Ligtenberg MJ, Kuiper RP, Geurts van Kessel A, et al.: EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. Fam Cancer 12 (2): 169-74, 2013. [PUBMED Abstract]
- Kempers MJ, Kuiper RP, Ockeloen CW, et al.: Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. Lancet Oncol 12 (1): 49-55, 2011. [PUBMED Abstract]
- Lynch HT, Riegert-Johnson DL, Snyder C, et al.: Lynch syndrome-associated extracolonic tumors are rare in two extended families with the same EPCAM deletion. Am J Gastroenterol 106 (10): 1829-36, 2011. [PUBMED Abstract]
- Dowty JG, Win AK, Buchanan DD, et al.: Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat 34 (3): 490-7, 2013. [PUBMED Abstract]
- International Mismatch Repair Consortium: Variation in the risk of colorectal cancer in families with Lynch syndrome: a retrospective cohort study. Lancet Oncol 22 (7): 1014-1022, 2021. [PUBMED Abstract]
- Desai DC, Lockman JC, Chadwick RB, et al.: Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet 37 (9): 646-52, 2000. [PUBMED Abstract]
- Nyström-Lahti M, Kristo P, Nicolaides NC, et al.: Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med 1 (11): 1203-6, 1995. [PUBMED Abstract]
- Moisio AL, Sistonen P, Weissenbach J, et al.: Age and origin of two common MLH1 mutations predisposing to hereditary colon cancer. Am J Hum Genet 59 (6): 1243-51, 1996. [PUBMED Abstract]
- Caluseriu O, Di Gregorio C, Lucci-Cordisco E, et al.: A founder MLH1 mutation in families from the districts of Modena and Reggio-Emilia in northern Italy with hereditary non-polyposis colorectal cancer associated with protein elongation and instability. J Med Genet 41 (3): e34, 2004. [PUBMED Abstract]
- Chan TL, Chan YW, Ho JW, et al.: MSH2 c.1452-1455delAATG is a founder mutation and an important cause of hereditary nonpolyposis colorectal cancer in the southern Chinese population. Am J Hum Genet 74 (5): 1035-42, 2004. [PUBMED Abstract]
- Clendenning M, Baze ME, Sun S, et al.: Origins and prevalence of the American Founder Mutation of MSH2. Cancer Res 68 (7): 2145-53, 2008. [PUBMED Abstract]
- Dominguez-Valentin M, Nilbert M, Wernhoff P, et al.: Mutation spectrum in South American Lynch syndrome families. Hered Cancer Clin Pract 11 (1): 18, 2013. [PUBMED Abstract]
- Cruz-Correa M, Diaz-Algorri Y, Pérez-Mayoral J, et al.: Clinical characterization and mutation spectrum in Caribbean Hispanic families with Lynch syndrome. Fam Cancer 14 (3): 415-25, 2015. [PUBMED Abstract]
- Sunga AY, Ricker C, Espenschied CR, et al.: Spectrum of mismatch repair gene mutations and clinical presentation of Hispanic individuals with Lynch syndrome. Cancer Genet 212-213: 1-7, 2017. [PUBMED Abstract]
- Ricker CN, Hanna DL, Peng C, et al.: DNA mismatch repair deficiency and hereditary syndromes in Latino patients with colorectal cancer. Cancer 123 (19): 3732-3743, 2017. [PUBMED Abstract]
- Guindalini RS, Win AK, Gulden C, et al.: Mutation spectrum and risk of colorectal cancer in African American families with Lynch syndrome. Gastroenterology 149 (6): 1446-53, 2015. [PUBMED Abstract]
- Parry S, Win AK, Parry B, et al.: Metachronous colorectal cancer risk for mismatch repair gene mutation carriers: the advantage of more extensive colon surgery. Gut 60 (7): 950-7, 2011. [PUBMED Abstract]
- Signoroni S, Piozzi GN, Ricci MT, et al.: Risk factors for metachronous colorectal cancer in Lynch syndrome patients: a registry-based observational mono-institutional study cohort. Int J Clin Oncol 25 (9): 1644-1652, 2020. [PUBMED Abstract]
- Watson P, Vasen HF, Mecklin JP, et al.: The risk of endometrial cancer in hereditary nonpolyposis colorectal cancer. Am J Med 96 (6): 516-20, 1994. [PUBMED Abstract]
- Watson P, Lynch HT: Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer 71 (3): 677-85, 1993. [PUBMED Abstract]
- Voskuil DW, Vasen HF, Kampman E, et al.: Colorectal cancer risk in HNPCC families: development during lifetime and in successive generations. National Collaborative Group on HNPCC. Int J Cancer 72 (2): 205-9, 1997. [PUBMED Abstract]
- Heinimann K, Müller H, Weber W, et al.: Disease expression in Swiss hereditary non-polyposis colorectal cancer (HNPCC) kindreds. Int J Cancer 74 (3): 281-5, 1997. [PUBMED Abstract]
- Lu KH, Dinh M, Kohlmann W, et al.: Gynecologic cancer as a "sentinel cancer" for women with hereditary nonpolyposis colorectal cancer syndrome. Obstet Gynecol 105 (3): 569-74, 2005. [PUBMED Abstract]
- Tan YY, McGaughran J, Ferguson K, et al.: Improving identification of lynch syndrome patients: a comparison of research data with clinical records. Int J Cancer 132 (12): 2876-83, 2013. [PUBMED Abstract]
- Win AK, Young JP, Lindor NM, et al.: Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol 30 (9): 958-64, 2012. [PUBMED Abstract]
- Win AK, Lindor NM, Winship I, et al.: Risks of colorectal and other cancers after endometrial cancer for women with Lynch syndrome. J Natl Cancer Inst 105 (4): 274-9, 2013. [PUBMED Abstract]
- Broaddus RR, Lynch HT, Chen LM, et al.: Pathologic features of endometrial carcinoma associated with HNPCC: a comparison with sporadic endometrial carcinoma. Cancer 106 (1): 87-94, 2006. [PUBMED Abstract]
- Vasen HF, Offerhaus GJ, den Hartog Jager FC, et al.: The tumour spectrum in hereditary non-polyposis colorectal cancer: a study of 24 kindreds in the Netherlands. Int J Cancer 46 (1): 31-4, 1990. [PUBMED Abstract]
- Aarnio M, Mecklin JP, Aaltonen LA, et al.: Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer 64 (6): 430-3, 1995. [PUBMED Abstract]
- Ketabi Z, Bartuma K, Bernstein I, et al.: Ovarian cancer linked to Lynch syndrome typically presents as early-onset, non-serous epithelial tumors. Gynecol Oncol 121 (3): 462-5, 2011. [PUBMED Abstract]
- Borelli I, Casalis Cavalchini GC, Del Peschio S, et al.: A founder MLH1 mutation in Lynch syndrome families from Piedmont, Italy, is associated with an increased risk of pancreatic tumours and diverse immunohistochemical patterns. Fam Cancer 13 (3): 401-13, 2014. [PUBMED Abstract]
- Raymond VM, Mukherjee B, Wang F, et al.: Elevated risk of prostate cancer among men with Lynch syndrome. J Clin Oncol 31 (14): 1713-8, 2013. [PUBMED Abstract]
- Raymond VM, Everett JN, Furtado LV, et al.: Adrenocortical carcinoma is a lynch syndrome-associated cancer. J Clin Oncol 31 (24): 3012-8, 2013. [PUBMED Abstract]
- Haraldsdottir S, Hampel H, Wei L, et al.: Prostate cancer incidence in males with Lynch syndrome. Genet Med 16 (7): 553-7, 2014. [PUBMED Abstract]
- Bapat B, Xia L, Madlensky L, et al.: The genetic basis of Muir-Torre syndrome includes the hMLH1 locus. Am J Hum Genet 59 (3): 736-9, 1996. [PUBMED Abstract]
- Lynch HT, Lynch PM, Pester J, et al.: The cancer family syndrome. Rare cutaneous phenotypic linkage of Torre's syndrome. Arch Intern Med 141 (5): 607-11, 1981. [PUBMED Abstract]
- Suspiro A, Fidalgo P, Cravo M, et al.: The Muir-Torre syndrome: a rare variant of hereditary nonpolyposis colorectal cancer associated with hMSH2 mutation. Am J Gastroenterol 93 (9): 1572-4, 1998. [PUBMED Abstract]
- Kruse R, Rütten A, Lamberti C, et al.: Muir-Torre phenotype has a frequency of DNA mismatch-repair-gene mutations similar to that in hereditary nonpolyposis colorectal cancer families defined by the Amsterdam criteria. Am J Hum Genet 63 (1): 63-70, 1998. [PUBMED Abstract]
- South CD, Hampel H, Comeras I, et al.: The frequency of Muir-Torre syndrome among Lynch syndrome families. J Natl Cancer Inst 100 (4): 277-81, 2008. [PUBMED Abstract]
- Kacerovska D, Cerna K, Martinek P, et al.: MSH6 mutation in a family affected by Muir-Torre syndrome. Am J Dermatopathol 34 (6): 648-52, 2012. [PUBMED Abstract]
- Tavakkol Z, Keller JJ, Furmanczyk PS, et al.: Germline mutation in MSH6 associated with multiple malignant neoplasms in a patient With Muir-Torre syndrome. J Clin Oncol 30 (22): e195-8, 2012. [PUBMED Abstract]
- Murphy HR, Armstrong R, Cairns D, et al.: Muir-Torre Syndrome: expanding the genotype and phenotype--a further family with a MSH6 mutation. Fam Cancer 7 (3): 255-7, 2008. [PUBMED Abstract]
- Arnold A, Payne S, Fisher S, et al.: An individual with Muir-Torre syndrome found to have a pathogenic MSH6 gene mutation. Fam Cancer 6 (3): 317-21, 2007. [PUBMED Abstract]
- Mangold E, Rahner N, Friedrichs N, et al.: MSH6 mutation in Muir-Torre syndrome: could this be a rare finding? Br J Dermatol 156 (1): 158-62, 2007. [PUBMED Abstract]
- Kastrinos F, Stoffel EM, Balmaña J, et al.: Phenotype comparison of MLH1 and MSH2 mutation carriers in a cohort of 1,914 individuals undergoing clinical genetic testing in the United States. Cancer Epidemiol Biomarkers Prev 17 (8): 2044-51, 2008. [PUBMED Abstract]
- Lamba AR, Moore AY, Moore T, et al.: Defective DNA mismatch repair activity is common in sebaceous neoplasms, and may be an ineffective approach to screen for Lynch syndrome. Fam Cancer 14 (2): 259-64, 2015. [PUBMED Abstract]
- Syngal S, Brand RE, Church JM, et al.: ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 110 (2): 223-62; quiz 263, 2015. [PUBMED Abstract]
- Howlader N, Noone AM, Krapcho M, et al., eds.: SEER Cancer Statistics Review (CSR) 1975-2014. National Cancer Institute. Also available online. Last accessed May 24, 2024.
- Jenkins MA, Baglietto L, Dowty JG, et al.: Cancer risks for mismatch repair gene mutation carriers: a population-based early onset case-family study. Clin Gastroenterol Hepatol 4 (4): 489-98, 2006. [PUBMED Abstract]
- Barrow E, Robinson L, Alduaij W, et al.: Cumulative lifetime incidence of extracolonic cancers in Lynch syndrome: a report of 121 families with proven mutations. Clin Genet 75 (2): 141-9, 2009. [PUBMED Abstract]
- Engel C, Loeffler M, Steinke V, et al.: Risks of less common cancers in proven mutation carriers with lynch syndrome. J Clin Oncol 30 (35): 4409-15, 2012. [PUBMED Abstract]
- Watson P, Vasen HF, Mecklin JP, et al.: The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int J Cancer 123 (2): 444-9, 2008. [PUBMED Abstract]
- Capelle LG, Van Grieken NC, Lingsma HF, et al.: Risk and epidemiological time trends of gastric cancer in Lynch syndrome carriers in the Netherlands. Gastroenterology 138 (2): 487-92, 2010. [PUBMED Abstract]
- Aarnio M, Sankila R, Pukkala E, et al.: Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer 81 (2): 214-8, 1999. [PUBMED Abstract]
- van der Post RS, Kiemeney LA, Ligtenberg MJ, et al.: Risk of urothelial bladder cancer in Lynch syndrome is increased, in particular among MSH2 mutation carriers. J Med Genet 47 (7): 464-70, 2010. [PUBMED Abstract]
- Cloyd JM, Chun YS, Ikoma N, et al.: Clinical and Genetic Implications of DNA Mismatch Repair Deficiency in Biliary Tract Cancers Associated with Lynch Syndrome. J Gastrointest Cancer 49 (1): 93-96, 2018. [PUBMED Abstract]
- Yang KY, Caughey AB, Little SE, et al.: A cost-effectiveness analysis of prophylactic surgery versus gynecologic surveillance for women from hereditary non-polyposis colorectal cancer (HNPCC) Families. Fam Cancer 10 (3): 535-43, 2011. [PUBMED Abstract]
- Ponti G, Losi L, Pedroni M, et al.: Value of MLH1 and MSH2 mutations in the appearance of Muir-Torre syndrome phenotype in HNPCC patients presenting sebaceous gland tumors or keratoacanthomas. J Invest Dermatol 126 (10): 2302-7, 2006. [PUBMED Abstract]
- Schwartz RA, Torre DP: The Muir-Torre syndrome: a 25-year retrospect. J Am Acad Dermatol 33 (1): 90-104, 1995. [PUBMED Abstract]
- Dunlop MG, Farrington SM, Carothers AD, et al.: Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet 6 (1): 105-10, 1997. [PUBMED Abstract]
- Kastrinos F, Mukherjee B, Tayob N, et al.: Risk of pancreatic cancer in families with Lynch syndrome. JAMA 302 (16): 1790-5, 2009. [PUBMED Abstract]
- Jensen UB, Sunde L, Timshel S, et al.: Mismatch repair defective breast cancer in the hereditary nonpolyposis colorectal cancer syndrome. Breast Cancer Res Treat 120 (3): 777-82, 2010. [PUBMED Abstract]
- Shanley S, Fung C, Milliken J, et al.: Breast cancer immunohistochemistry can be useful in triage of some HNPCC families. Fam Cancer 8 (3): 251-5, 2009. [PUBMED Abstract]
- Walsh MD, Buchanan DD, Cummings MC, et al.: Lynch syndrome-associated breast cancers: clinicopathologic characteristics of a case series from the colon cancer family registry. Clin Cancer Res 16 (7): 2214-24, 2010. [PUBMED Abstract]
- Buerki N, Gautier L, Kovac M, et al.: Evidence for breast cancer as an integral part of Lynch syndrome. Genes Chromosomes Cancer 51 (1): 83-91, 2012. [PUBMED Abstract]
- Win AK, Lindor NM, Young JP, et al.: Risks of primary extracolonic cancers following colorectal cancer in lynch syndrome. J Natl Cancer Inst 104 (18): 1363-72, 2012. [PUBMED Abstract]
- Harkness EF, Barrow E, Newton K, et al.: Lynch syndrome caused by MLH1 mutations is associated with an increased risk of breast cancer: a cohort study. J Med Genet 52 (8): 553-6, 2015. [PUBMED Abstract]
- Win AK, Lindor NM, Jenkins MA: Risk of breast cancer in Lynch syndrome: a systematic review. Breast Cancer Res 15 (2): R27, 2013. [PUBMED Abstract]
- Goldberg M, Bell K, Aronson M, et al.: Association between the Lynch syndrome gene MSH2 and breast cancer susceptibility in a Canadian familial cancer registry. J Med Genet 54 (11): 742-746, 2017. [PUBMED Abstract]
- Lu HM, Li S, Black MH, et al.: Association of Breast and Ovarian Cancers With Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol 5 (1): 51-57, 2019. [PUBMED Abstract]
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic. Version 1.2021. Plymouth Meeting, Pa: National Comprehensive Cancer Network, 2020. Available online with free registration. Last accessed May 24, 2024.
- Ryan S, Jenkins MA, Win AK: Risk of prostate cancer in Lynch syndrome: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev 23 (3): 437-49, 2014. [PUBMED Abstract]
- Pritchard CC, Mateo J, Walsh MF, et al.: Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 375 (5): 443-53, 2016. [PUBMED Abstract]
- De Jong AE, Morreau H, Van Puijenbroek M, et al.: The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology 126 (1): 42-8, 2004. [PUBMED Abstract]
- Johnson PM, Gallinger S, McLeod RS: Surveillance colonoscopy in individuals at risk for hereditary nonpolyposis colorectal cancer: an evidence-based review. Dis Colon Rectum 49 (1): 80-93; discussion 94-5, 2006. [PUBMED Abstract]
- Lindor NM, Petersen GM, Hadley DW, et al.: Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA 296 (12): 1507-17, 2006. [PUBMED Abstract]
- Reitmair AH, Cai JC, Bjerknes M, et al.: MSH2 deficiency contributes to accelerated APC-mediated intestinal tumorigenesis. Cancer Res 56 (13): 2922-6, 1996. [PUBMED Abstract]
- Järvinen HJ, Aarnio M, Mustonen H, et al.: Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 118 (5): 829-34, 2000. [PUBMED Abstract]
- Järvinen HJ, Mecklin JP, Sistonen P: Screening reduces colorectal cancer rate in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 108 (5): 1405-11, 1995. [PUBMED Abstract]
- Engel C, Rahner N, Schulmann K, et al.: Efficacy of annual colonoscopic surveillance in individuals with hereditary nonpolyposis colorectal cancer. Clin Gastroenterol Hepatol 8 (2): 174-82, 2010. [PUBMED Abstract]
- Vasen HF, Abdirahman M, Brohet R, et al.: One to 2-year surveillance intervals reduce risk of colorectal cancer in families with Lynch syndrome. Gastroenterology 138 (7): 2300-6, 2010. [PUBMED Abstract]
- Järvinen HJ, Renkonen-Sinisalo L, Aktán-Collán K, et al.: Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J Clin Oncol 27 (28): 4793-7, 2009. [PUBMED Abstract]
- Lindberg LJ, Wegen-Haitsma W, Ladelund S, et al.: Risk of multiple colorectal cancer development depends on age and subgroup in individuals with hereditary predisposition. Fam Cancer 18 (2): 183-191, 2019. [PUBMED Abstract]
- Hurlstone DP, Karajeh M, Cross SS, et al.: The role of high-magnification-chromoscopic colonoscopy in hereditary nonpolyposis colorectal cancer screening: a prospective "back-to-back" endoscopic study. Am J Gastroenterol 100 (10): 2167-73, 2005. [PUBMED Abstract]
- Lecomte T, Cellier C, Meatchi T, et al.: Chromoendoscopic colonoscopy for detecting preneoplastic lesions in hereditary nonpolyposis colorectal cancer syndrome. Clin Gastroenterol Hepatol 3 (9): 897-902, 2005. [PUBMED Abstract]
- Müller A, Beckmann C, Westphal G, et al.: Prevalence of the mismatch-repair-deficient phenotype in colonic adenomas arising in HNPCC patients: results of a 5-year follow-up study. Int J Colorectal Dis 21 (7): 632-41, 2006. [PUBMED Abstract]
- Yurgelun MB, Goel A, Hornick JL, et al.: Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev Res (Phila) 5 (4): 574-82, 2012. [PUBMED Abstract]
- Monahan KJ, Bradshaw N, Dolwani S, et al.: Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG). Gut 69 (3): 411-444, 2020. [PUBMED Abstract]
- Seppälä TT, Latchford A, Negoi I, et al.: European guidelines from the EHTG and ESCP for Lynch syndrome: an updated third edition of the Mallorca guidelines based on gene and gender. Br J Surg 108 (5): 484-498, 2021. [PUBMED Abstract]
- Stjepanovic N, Moreira L, Carneiro F, et al.: Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 30 (10): 1558-1571, 2019. [PUBMED Abstract]
- Hendriks YM, de Jong AE, Morreau H, et al.: Diagnostic approach and management of Lynch syndrome (hereditary nonpolyposis colorectal carcinoma): a guide for clinicians. CA Cancer J Clin 56 (4): 213-25, 2006 Jul-Aug. [PUBMED Abstract]
- Dove-Edwin I, Boks D, Goff S, et al.: The outcome of endometrial carcinoma surveillance by ultrasound scan in women at risk of hereditary nonpolyposis colorectal carcinoma and familial colorectal carcinoma. Cancer 94 (6): 1708-12, 2002. [PUBMED Abstract]
- Rijcken FE, Mourits MJ, Kleibeuker JH, et al.: Gynecologic screening in hereditary nonpolyposis colorectal cancer. Gynecol Oncol 91 (1): 74-80, 2003. [PUBMED Abstract]
- Renkonen-Sinisalo L, Bützow R, Leminen A, et al.: Surveillance for endometrial cancer in hereditary nonpolyposis colorectal cancer syndrome. Int J Cancer 120 (4): 821-4, 2007. [PUBMED Abstract]
- Yang K, Allen B, Conrad P, et al.: Awareness of gynecologic surveillance in women from hereditary non-polyposis colorectal cancer families. Fam Cancer 5 (4): 405-9, 2006. [PUBMED Abstract]
- Collins VR, Meiser B, Ukoumunne OC, et al.: The impact of predictive genetic testing for hereditary nonpolyposis colorectal cancer: three years after testing. Genet Med 9 (5): 290-7, 2007. [PUBMED Abstract]
- Schmeler KM, Lynch HT, Chen LM, et al.: Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N Engl J Med 354 (3): 261-9, 2006. [PUBMED Abstract]
- Kwon JS, Sun CC, Peterson SK, et al.: Cost-effectiveness analysis of prevention strategies for gynecologic cancers in Lynch syndrome. Cancer 113 (2): 326-35, 2008. [PUBMED Abstract]
- Aarnio M, Salovaara R, Aaltonen LA, et al.: Features of gastric cancer in hereditary non-polyposis colorectal cancer syndrome. Int J Cancer 74 (5): 551-5, 1997. [PUBMED Abstract]
- Farha N, Hrabe J, Sleiman J, et al.: Clinically actionable findings on surveillance EGD in asymptomatic patients with Lynch syndrome. Gastrointest Endosc 95 (1): 105-114, 2022. [PUBMED Abstract]
- Canto MI, Harinck F, Hruban RH, et al.: International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 62 (3): 339-47, 2013. [PUBMED Abstract]
- Burn J, Gerdes AM, Macrae F, et al.: Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet 378 (9809): 2081-7, 2011. [PUBMED Abstract]
- Burn J, Bishop DT, Mecklin JP, et al.: Effect of aspirin or resistant starch on colorectal neoplasia in the Lynch syndrome. N Engl J Med 359 (24): 2567-78, 2008. [PUBMED Abstract]
- Burn J, Sheth H, Elliott F, et al.: Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: a double-blind, randomised, placebo-controlled trial. Lancet 395 (10240): 1855-1863, 2020. [PUBMED Abstract]
- Mathers JC, Elliott F, Macrae F, et al.: Cancer Prevention with Resistant Starch in Lynch Syndrome Patients in the CAPP2-Randomized Placebo Controlled Trial: Planned 10-Year Follow-up. Cancer Prev Res (Phila) 15 (9): 623-634, 2022. [PUBMED Abstract]
- Yurgelun MB, Chan AT: Aspirin for Lynch syndrome: a legacy of prevention. Lancet 395 (10240): 1817-1818, 2020. [PUBMED Abstract]
- Movahedi M, Bishop DT, Macrae F, et al.: Obesity, Aspirin, and Risk of Colorectal Cancer in Carriers of Hereditary Colorectal Cancer: A Prospective Investigation in the CAPP2 Study. J Clin Oncol 33 (31): 3591-7, 2015. [PUBMED Abstract]
- Burn J, Mathers JC, Bishop DT: Chemoprevention in Lynch syndrome. Fam Cancer 12 (4): 707-18, 2013. [PUBMED Abstract]
- Ait Ouakrim D, Dashti SG, Chau R, et al.: Aspirin, Ibuprofen, and the Risk of Colorectal Cancer in Lynch Syndrome. J Natl Cancer Inst 107 (9): , 2015. [PUBMED Abstract]
- Reyes-Uribe L, Wu W, Gelincik O, et al.: Naproxen chemoprevention promotes immune activation in Lynch syndrome colorectal mucosa. Gut 70 (3): 555-566, 2021. [PUBMED Abstract]
- de Vos tot Nederveen Cappel WH, Buskens E, van Duijvendijk P, et al.: Decision analysis in the surgical treatment of colorectal cancer due to a mismatch repair gene defect. Gut 52 (12): 1752-5, 2003. [PUBMED Abstract]
- Natarajan N, Watson P, Silva-Lopez E, et al.: Comparison of extended colectomy and limited resection in patients with Lynch syndrome. Dis Colon Rectum 53 (1): 77-82, 2010. [PUBMED Abstract]
- Maeda T, Cannom RR, Beart RW, et al.: Decision model of segmental compared with total abdominal colectomy for colon cancer in hereditary nonpolyposis colorectal cancer. J Clin Oncol 28 (7): 1175-80, 2010. [PUBMED Abstract]
- Hiatt MJ, Casey MJ, Lynch HT, et al.: Efficacy of proximal colectomy for surgical management of right-sided first colorectal cancer in Lynch Syndrome mutation carriers. Am J Surg 216 (1): 99-105, 2018. [PUBMED Abstract]
- Rodríguez-Bigas MA, Vasen HF, Pekka-Mecklin J, et al.: Rectal cancer risk in hereditary nonpolyposis colorectal cancer after abdominal colectomy. International Collaborative Group on HNPCC. Ann Surg 225 (2): 202-7, 1997. [PUBMED Abstract]
- de Rosa N, Rodriguez-Bigas MA, Chang GJ, et al.: DNA Mismatch Repair Deficiency in Rectal Cancer: Benchmarking Its Impact on Prognosis, Neoadjuvant Response Prediction, and Clinical Cancer Genetics. J Clin Oncol 34 (25): 3039-46, 2016. [PUBMED Abstract]
- Lee JS, Petrelli NJ, Rodriguez-Bigas MA: Rectal cancer in hereditary nonpolyposis colorectal cancer. Am J Surg 181 (3): 207-10, 2001. [PUBMED Abstract]
- Kalady MF, Lipman J, McGannon E, et al.: Risk of colonic neoplasia after proctectomy for rectal cancer in hereditary nonpolyposis colorectal cancer. Ann Surg 255 (6): 1121-5, 2012. [PUBMED Abstract]
- Olsen KØ, Juul S, Bülow S, et al.: Female fecundity before and after operation for familial adenomatous polyposis. Br J Surg 90 (2): 227-31, 2003. [PUBMED Abstract]
- Nieuwenhuis MH, Douma KF, Bleiker EM, et al.: Female fertility after colorectal surgery for familial adenomatous polyposis: a nationwide cross-sectional study. Ann Surg 252 (2): 341-4, 2010. [PUBMED Abstract]
- Guillem JG, Wood WC, Moley JF, et al.: ASCO/SSO review of current role of risk-reducing surgery in common hereditary cancer syndromes. J Clin Oncol 24 (28): 4642-60, 2006. [PUBMED Abstract]
- Vasen HF, Blanco I, Aktan-Collan K, et al.: Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 62 (6): 812-23, 2013. [PUBMED Abstract]
- Rodriguez-Bigas MA, Möeslein G: Surgical treatment of hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome). Fam Cancer 12 (2): 295-300, 2013. [PUBMED Abstract]
- Samowitz WS, Curtin K, Ma KN, et al.: Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev 10 (9): 917-23, 2001. [PUBMED Abstract]
- Koopman M, Kortman GA, Mekenkamp L, et al.: Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 100 (2): 266-73, 2009. [PUBMED Abstract]
- Popat S, Hubner R, Houlston RS: Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23 (3): 609-18, 2005. [PUBMED Abstract]
- Hutchins G, Southward K, Handley K, et al.: Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol 29 (10): 1261-70, 2011. [PUBMED Abstract]
- Roth AD, Tejpar S, Delorenzi M, et al.: Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 28 (3): 466-74, 2010. [PUBMED Abstract]
- Liu GC, Liu RY, Yan JP, et al.: The Heterogeneity Between Lynch-Associated and Sporadic MMR Deficiency in Colorectal Cancers. J Natl Cancer Inst 110 (9): 975-984, 2018. [PUBMED Abstract]
- Boland CR, Goel A: Microsatellite instability in colorectal cancer. Gastroenterology 138 (6): 2073-2087.e3, 2010. [PUBMED Abstract]
- Hawn MT, Umar A, Carethers JM, et al.: Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res 55 (17): 3721-5, 1995. [PUBMED Abstract]
- Carethers JM, Hawn MT, Chauhan DP, et al.: Competency in mismatch repair prohibits clonal expansion of cancer cells treated with N-methyl-N'-nitro-N-nitrosoguanidine. J Clin Invest 98 (1): 199-206, 1996. [PUBMED Abstract]
- Aebi S, Kurdi-Haidar B, Gordon R, et al.: Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res 56 (13): 3087-90, 1996. [PUBMED Abstract]
- Carethers JM, Chauhan DP, Fink D, et al.: Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology 117 (1): 123-31, 1999. [PUBMED Abstract]
- Elsaleh H, Joseph D, Grieu F, et al.: Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet 355 (9217): 1745-50, 2000. [PUBMED Abstract]
- Ribic CM, Sargent DJ, Moore MJ, et al.: Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 349 (3): 247-57, 2003. [PUBMED Abstract]
- Sinicrope FA, Foster NR, Thibodeau SN, et al.: DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst 103 (11): 863-75, 2011. [PUBMED Abstract]
- Fink D, Nebel S, Aebi S, et al.: The role of DNA mismatch repair in platinum drug resistance. Cancer Res 56 (21): 4881-6, 1996. [PUBMED Abstract]
- Tougeron D, Mouillet G, Trouilloud I, et al.: Efficacy of Adjuvant Chemotherapy in Colon Cancer With Microsatellite Instability: A Large Multicenter AGEO Study. J Natl Cancer Inst 108 (7): , 2016. [PUBMED Abstract]
- Kim JE, Hong YS, Kim HJ, et al.: Microsatellite Instability was not Associated with Survival in Stage III Colon Cancer Treated with Adjuvant Chemotherapy of Oxaliplatin and Infusional 5-Fluorouracil and Leucovorin (FOLFOX). Ann Surg Oncol 24 (5): 1289-1294, 2017. [PUBMED Abstract]
- Oh SY, Kim DY, Kim YB, et al.: Oncologic outcomes after adjuvant chemotherapy using FOLFOX in MSI-H sporadic stage III colon cancer. World J Surg 37 (10): 2497-503, 2013. [PUBMED Abstract]
- Le DT, Uram JN, Wang H, et al.: PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372 (26): 2509-20, 2015. [PUBMED Abstract]
- Overman MJ, McDermott R, Leach JL, et al.: Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 18 (9): 1182-1191, 2017. [PUBMED Abstract]
- André T, Shiu KK, Kim TW, et al.: Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med 383 (23): 2207-2218, 2020. [PUBMED Abstract]
- Diaz LA, Shiu KK, Kim TW, et al.: Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol 23 (5): 659-670, 2022. [PUBMED Abstract]
- Overman MJ, Lonardi S, Wong KYM, et al.: Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol 36 (8): 773-779, 2018. [PUBMED Abstract]
- Ludford K, Cohen R, Svrcek M, et al.: Pathological Tumor Response Following Immune Checkpoint Blockade for Deficient Mismatch Repair Advanced Colorectal Cancer. J Natl Cancer Inst 113 (2): 208-211, 2021. [PUBMED Abstract]
- Zhou XP, Waite KA, Pilarski R, et al.: Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet 73 (2): 404-11, 2003. [PUBMED Abstract]
- Mester J, Eng C: When overgrowth bumps into cancer: the PTEN-opathies. Am J Med Genet C Semin Med Genet 163C (2): 114-21, 2013. [PUBMED Abstract]
- Eng C: PTEN: one gene, many syndromes. Hum Mutat 22 (3): 183-98, 2003. [PUBMED Abstract]
- Marsh DJ, Kum JB, Lunetta KL, et al.: PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8 (8): 1461-72, 1999. [PUBMED Abstract]
- Pilarski R, Eng C: Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet 41 (5): 323-6, 2004. [PUBMED Abstract]
- Eng C: PTEN Hamartoma Tumor Syndrome (PHTS). In: Adam MP, Feldman J, Mirzaa GM, et al., eds.: GeneReviews. University of Washington, Seattle, 1993-2024, pp. Available online. Last accessed May 8, 2025.
- Pilarski R, Burt R, Kohlman W, et al.: Cowden syndrome and the PTEN hamartoma tumor syndrome: systematic review and revised diagnostic criteria. J Natl Cancer Inst 105 (21): 1607-16, 2013. [PUBMED Abstract]
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic. Version 2.2022. Plymouth Meeting, Pa: National Comprehensive Cancer Network, 2022. Available online with free registration. Last accessed May 24, 2024.
- Hampel H, Bennett RL, Buchanan A, et al.: A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med 17 (1): 70-87, 2015. [PUBMED Abstract]
- Ngeow J, Liu C, Zhou K, et al.: Detecting Germline PTEN Mutations Among At-Risk Patients With Cancer: An Age- and Sex-Specific Cost-Effectiveness Analysis. J Clin Oncol 33 (23): 2537-44, 2015. [PUBMED Abstract]
- Tan MH, Mester JL, Ngeow J, et al.: Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 18 (2): 400-7, 2012. [PUBMED Abstract]
- Bubien V, Bonnet F, Brouste V, et al.: High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet 50 (4): 255-63, 2013. [PUBMED Abstract]
- Heald B, Mester J, Rybicki L, et al.: Frequent gastrointestinal polyps and colorectal adenocarcinomas in a prospective series of PTEN mutation carriers. Gastroenterology 139 (6): 1927-33, 2010. [PUBMED Abstract]
- Peutz JL: Very remarkable case of familial polyposis of mucous membrane of intestinal tract and nasopharynx accompanied by peculiar pigmentations of skin and mucous membrane. Ned Tijdschr Geneeskd 10: 134-146, 1921.
- Jeghers H, McKusick VA, Katz KH: Generalized intestinal polyposis and melanin spots of the oral mucosa, lips and digits; a syndrome of diagnostic significance. N Engl J Med 241 (26): 1031-6, 1949. [PUBMED Abstract]
- Spigelman AD, Murday V, Phillips RK: Cancer and the Peutz-Jeghers syndrome. Gut 30 (11): 1588-90, 1989. [PUBMED Abstract]
- Aretz S, Stienen D, Uhlhaas S, et al.: High proportion of large genomic STK11 deletions in Peutz-Jeghers syndrome. Hum Mutat 26 (6): 513-9, 2005. [PUBMED Abstract]
- Hemminki A, Markie D, Tomlinson I, et al.: A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 391 (6663): 184-7, 1998. [PUBMED Abstract]
- Jenne DE, Reimann H, Nezu J, et al.: Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet 18 (1): 38-43, 1998. [PUBMED Abstract]
- Boudeau J, Kieloch A, Alessi DR, et al.: Functional analysis of LKB1/STK11 mutants and two aberrant isoforms found in Peutz-Jeghers Syndrome patients. Hum Mutat 21 (2): 172, 2003. [PUBMED Abstract]
- Lim W, Hearle N, Shah B, et al.: Further observations on LKB1/STK11 status and cancer risk in Peutz-Jeghers syndrome. Br J Cancer 89 (2): 308-13, 2003. [PUBMED Abstract]
- Giardiello FM, Brensinger JD, Tersmette AC, et al.: Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology 119 (6): 1447-53, 2000. [PUBMED Abstract]
- Lim W, Olschwang S, Keller JJ, et al.: Relative frequency and morphology of cancers in STK11 mutation carriers. Gastroenterology 126 (7): 1788-94, 2004. [PUBMED Abstract]
- van Lier MG, Wagner A, Mathus-Vliegen EM, et al.: High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol 105 (6): 1258-64; author reply 1265, 2010. [PUBMED Abstract]
- Srivatsa PJ, Keeney GL, Podratz KC: Disseminated cervical adenoma malignum and bilateral ovarian sex cord tumors with annular tubules associated with Peutz-Jeghers syndrome. Gynecol Oncol 53 (2): 256-64, 1994. [PUBMED Abstract]
- Scully RE: Sex cord tumor with annular tubules a distinctive ovarian tumor of the Peutz-Jeghers syndrome. Cancer 25 (5): 1107-21, 1970. [PUBMED Abstract]
- Westerman AM, Entius MM, de Baar E, et al.: Peutz-Jeghers syndrome: 78-year follow-up of the original family. Lancet 353 (9160): 1211-5, 1999. [PUBMED Abstract]
- Mehenni H, Resta N, Park JG, et al.: Cancer risks in LKB1 germline mutation carriers. Gut 55 (7): 984-90, 2006. [PUBMED Abstract]
- Gruber SB, Entius MM, Petersen GM, et al.: Pathogenesis of adenocarcinoma in Peutz-Jeghers syndrome. Cancer Res 58 (23): 5267-70, 1998. [PUBMED Abstract]
- Wang ZJ, Ellis I, Zauber P, et al.: Allelic imbalance at the LKB1 (STK11) locus in tumours from patients with Peutz-Jeghers' syndrome provides evidence for a hamartoma-(adenoma)-carcinoma sequence. J Pathol 188 (1): 9-13, 1999. [PUBMED Abstract]
- Miyoshi H, Nakau M, Ishikawa TO, et al.: Gastrointestinal hamartomatous polyposis in Lkb1 heterozygous knockout mice. Cancer Res 62 (8): 2261-6, 2002. [PUBMED Abstract]
- Nakau M, Miyoshi H, Seldin MF, et al.: Hepatocellular carcinoma caused by loss of heterozygosity in Lkb1 gene knockout mice. Cancer Res 62 (16): 4549-53, 2002. [PUBMED Abstract]
- Takeda H, Miyoshi H, Kojima Y, et al.: Accelerated onsets of gastric hamartomas and hepatic adenomas/carcinomas in Lkb1+/-p53-/- compound mutant mice. Oncogene 25 (12): 1816-20, 2006. [PUBMED Abstract]
- Amos CI, Keitheri-Cheteri MB, Sabripour M, et al.: Genotype-phenotype correlations in Peutz-Jeghers syndrome. J Med Genet 41 (5): 327-33, 2004. [PUBMED Abstract]
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal. Version 2.2023 . Plymouth Meeting, PA: National Comprehensive Cancer Network, 2023. Available with free registration. Last accessed March 13, 2024.
- Boland CR, Idos GE, Durno C, et al.: Diagnosis and Management of Cancer Risk in the Gastrointestinal Hamartomatous Polyposis Syndromes: Recommendations From the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 162 (7): 2063-2085, 2022. [PUBMED Abstract]
- Urquhart P, Grimpen F, Lim GJ, et al.: Capsule endoscopy versus magnetic resonance enterography for the detection of small bowel polyps in Peutz-Jeghers syndrome. Fam Cancer 13 (2): 249-55, 2014. [PUBMED Abstract]
- Latchford AR, Neale K, Phillips RK, et al.: Juvenile polyposis syndrome: a study of genotype, phenotype, and long-term outcome. Dis Colon Rectum 55 (10): 1038-43, 2012. [PUBMED Abstract]
- Veale AM, McColl I, Bussey HJ, et al.: Juvenile polyposis coli. J Med Genet 3 (1): 5-16, 1966. [PUBMED Abstract]
- Chow E, Macrae F: A review of juvenile polyposis syndrome. J Gastroenterol Hepatol 20 (11): 1634-40, 2005. [PUBMED Abstract]
- Jass JR, Williams CB, Bussey HJ, et al.: Juvenile polyposis--a precancerous condition. Histopathology 13 (6): 619-30, 1988. [PUBMED Abstract]
- Howe JR, Bair JL, Sayed MG, et al.: Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet 28 (2): 184-7, 2001. [PUBMED Abstract]
- Zhou XP, Woodford-Richens K, Lehtonen R, et al.: Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am J Hum Genet 69 (4): 704-11, 2001. [PUBMED Abstract]
- Aytac E, Sulu B, Heald B, et al.: Genotype-defined cancer risk in juvenile polyposis syndrome. Br J Surg 102 (1): 114-8, 2015. [PUBMED Abstract]
- Brosens LA, van Hattem A, Hylind LM, et al.: Risk of colorectal cancer in juvenile polyposis. Gut 56 (7): 965-7, 2007. [PUBMED Abstract]
- MacFarland SP, Ebrahimzadeh JE, Zelley K, et al.: Phenotypic Differences in Juvenile Polyposis Syndrome With or Without a Disease-causing SMAD4/BMPR1A Variant. Cancer Prev Res (Phila) 14 (2): 215-222, 2021. [PUBMED Abstract]
- Gallione CJ, Repetto GM, Legius E, et al.: A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 363 (9412): 852-9, 2004. [PUBMED Abstract]
- Rosner G, Petel-Galil Y, Laish I, et al.: Adenomatous Polyposis Phenotype in BMPR1A and SMAD4 Variant Carriers. Clin Transl Gastroenterol 13 (10): e00527, 2022. [PUBMED Abstract]
- Aretz S, Stienen D, Uhlhaas S, et al.: High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome. J Med Genet 44 (11): 702-9, 2007. [PUBMED Abstract]
- O'Malley M, LaGuardia L, Kalady MF, et al.: The prevalence of hereditary hemorrhagic telangiectasia in juvenile polyposis syndrome. Dis Colon Rectum 55 (8): 886-92, 2012. [PUBMED Abstract]
- Dahdaleh FS, Carr JC, Calva D, et al.: Juvenile polyposis and other intestinal polyposis syndromes with microdeletions of chromosome 10q22-23. Clin Genet 81 (2): 110-6, 2012. [PUBMED Abstract]
- Stanich PP, Pearlman R, Hinton A, et al.: Prevalence of Germline Mutations in Polyposis and Colorectal Cancer-Associated Genes in Patients With Multiple Colorectal Polyps. Clin Gastroenterol Hepatol 17 (10): 2008-2015.e3, 2019. [PUBMED Abstract]
- Yurgelun MB, Hornick JL, Curry VK, et al.: Therapy-associated polyposis as a late sequela of cancer treatment. Clin Gastroenterol Hepatol 12 (6): 1046-50, 2014. [PUBMED Abstract]
- Rigter LS, Kallenberg FG, Bastiaansen B, et al.: A case series of intestinal adenomatous polyposis of unidentified etiology; a late effect of irradiation? BMC Cancer 16 (1): 862, 2016. [PUBMED Abstract]
- Rigter LS, Spaander MCW, Aleman BMP, et al.: High prevalence of advanced colorectal neoplasia and serrated polyposis syndrome in Hodgkin lymphoma survivors. Cancer 125 (6): 990-999, 2019. [PUBMED Abstract]
- Palles C, Cazier JB, Howarth KM, et al.: Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45 (2): 136-44, 2013. [PUBMED Abstract]
- Briggs S, Tomlinson I: Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 230 (2): 148-53, 2013. [PUBMED Abstract]
- Cancer Genome Atlas Network: Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 (7407): 330-7, 2012. [PUBMED Abstract]
- Wang F, Zhao Q, Wang YN, et al.: Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol 5 (10): 1504-1506, 2019. [PUBMED Abstract]
- Mur P, García-Mulero S, Del Valle J, et al.: Role of POLE and POLD1 in familial cancer. Genet Med 22 (12): 2089-2100, 2020. [PUBMED Abstract]
- Buchanan DD, Stewart JR, Clendenning M, et al.: Risk of colorectal cancer for carriers of a germ-line mutation in POLE or POLD1. Genet Med 20 (8): 890-895, 2018. [PUBMED Abstract]
- Hamzaoui N, Alarcon F, Leulliot N, et al.: Genetic, structural, and functional characterization of POLE polymerase proofreading variants allows cancer risk prediction. Genet Med 22 (9): 1533-1541, 2020. [PUBMED Abstract]
- Weren RD, Ligtenberg MJ, Kets CM, et al.: A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet 47 (6): 668-71, 2015. [PUBMED Abstract]
- Broderick P, Dobbins SE, Chubb D, et al.: Validation of Recently Proposed Colorectal Cancer Susceptibility Gene Variants in an Analysis of Families and Patients-a Systematic Review. Gastroenterology 152 (1): 75-77.e4, 2017. [PUBMED Abstract]
- Beck SH, Jelsig AM, Yassin HM, et al.: Intestinal and extraintestinal neoplasms in patients with NTHL1 tumor syndrome: a systematic review. Fam Cancer 21 (4): 453-462, 2022. [PUBMED Abstract]
- Chan JM, Clendenning M, Joseland S, et al.: Rare germline variants in the AXIN2 gene in families with colonic polyposis and colorectal cancer. Fam Cancer 21 (4): 399-413, 2022. [PUBMED Abstract]
- Leclerc J, Beaumont M, Vibert R, et al.: AXIN2 germline testing in a French cohort validates pathogenic variants as a rare cause of predisposition to colorectal polyposis and cancer. Genes Chromosomes Cancer 62 (4): 210-222, 2023. [PUBMED Abstract]
- Lammi L, Arte S, Somer M, et al.: Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet 74 (5): 1043-50, 2004. [PUBMED Abstract]
- Marvin ML, Mazzoni SM, Herron CM, et al.: AXIN2-associated autosomal dominant ectodermal dysplasia and neoplastic syndrome. Am J Med Genet A 155A (4): 898-902, 2011. [PUBMED Abstract]
- Rivera B, Perea J, Sánchez E, et al.: A novel AXIN2 germline variant associated with attenuated FAP without signs of oligondontia or ectodermal dysplasia. Eur J Hum Genet 22 (3): 423-6, 2014. [PUBMED Abstract]
- Jaeger E, Leedham S, Lewis A, et al.: Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet 44 (6): 699-703, 2012. [PUBMED Abstract]
- Meijers-Heijboer H, Wijnen J, Vasen H, et al.: The CHEK2 1100delC mutation identifies families with a hereditary breast and colorectal cancer phenotype. Am J Hum Genet 72 (5): 1308-14, 2003. [PUBMED Abstract]
- Cybulski C, Górski B, Huzarski T, et al.: CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 75 (6): 1131-5, 2004. [PUBMED Abstract]
- de Jong MM, Nolte IM, Te Meerman GJ, et al.: Colorectal cancer and the CHEK2 1100delC mutation. Genes Chromosomes Cancer 43 (4): 377-82, 2005. [PUBMED Abstract]
- Uson PLS, Riegert-Johnson D, Boardman L, et al.: Germline Cancer Susceptibility Gene Testing in Unselected Patients With Colorectal Adenocarcinoma: A Multicenter Prospective Study. Clin Gastroenterol Hepatol 20 (3): e508-e528, 2022. [PUBMED Abstract]
- Breen KE, Katona BW, Catchings A, et al.: An updated counseling framework for moderate-penetrance colorectal cancer susceptibility genes. Genet Med 24 (12): 2587-2590, 2022. [PUBMED Abstract]
- Bychkovsky BL, Agaoglu NB, Horton C, et al.: Differences in Cancer Phenotypes Among Frequent CHEK2 Variants and Implications for Clinical Care-Checking CHEK2. JAMA Oncol 8 (11): 1598-1606, 2022. [PUBMED Abstract]
- Mundt E, Mabey B, Rainville I, et al.: Breast and colorectal cancer risks among over 6,000 CHEK2 pathogenic variant carriers: A comparison of missense versus truncating variants. Cancer Genet 278-279: 84-90, 2023. [PUBMED Abstract]
- Hazewinkel Y, López-Cerón M, East JE, et al.: Endoscopic features of sessile serrated adenomas: validation by international experts using high-resolution white-light endoscopy and narrow-band imaging. Gastrointest Endosc 77 (6): 916-24, 2013. [PUBMED Abstract]
- Guarinos C, Juárez M, Egoavil C, et al.: Prevalence and characteristics of MUTYH-associated polyposis in patients with multiple adenomatous and serrated polyps. Clin Cancer Res 20 (5): 1158-68, 2014. [PUBMED Abstract]
- Crockett SD, Snover DC, Ahnen DJ, et al.: Sessile serrated adenomas: an evidence-based guide to management. Clin Gastroenterol Hepatol 13 (1): 11-26.e1, 2015. [PUBMED Abstract]
- Boparai KS, Mathus-Vliegen EM, Koornstra JJ, et al.: Increased colorectal cancer risk during follow-up in patients with hyperplastic polyposis syndrome: a multicentre cohort study. Gut 59 (8): 1094-100, 2010. [PUBMED Abstract]
- Clendenning M, Young JP, Walsh MD, et al.: Germline Mutations in the Polyposis-Associated Genes BMPR1A, SMAD4, PTEN, MUTYH and GREM1 Are Not Common in Individuals with Serrated Polyposis Syndrome. PLoS One 8 (6): e66705, 2013. [PUBMED Abstract]
- Yan HHN, Lai JCW, Ho SL, et al.: RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut 66 (9): 1645-1656, 2017. [PUBMED Abstract]
- Quintana I, Mejías-Luque R, Terradas M, et al.: Evidence suggests that germline RNF43 mutations are a rare cause of serrated polyposis. Gut 67 (12): 2230-2232, 2018. [PUBMED Abstract]
- Taupin D, Lam W, Rangiah D, et al.: A deleterious RNF43 germline mutation in a severely affected serrated polyposis kindred. Hum Genome Var 2: 15013, 2015. [PUBMED Abstract]
- Dekker E, Bleijenberg A, Balaguer F, et al.: Update on the World Health Organization Criteria for Diagnosis of Serrated Polyposis Syndrome. Gastroenterology 158 (6): 1520-1523, 2020. [PUBMED Abstract]
- Leggett BA, Devereaux B, Biden K, et al.: Hyperplastic polyposis: association with colorectal cancer. Am J Surg Pathol 25 (2): 177-84, 2001. [PUBMED Abstract]
- Rashid A, Houlihan PS, Booker S, et al.: Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology 119 (2): 323-32, 2000. [PUBMED Abstract]
- Place RJ, Simmang CL: Hyperplastic-adenomatous polyposis syndrome. J Am Coll Surg 188 (5): 503-7, 1999. [PUBMED Abstract]
- Hyman NH, Anderson P, Blasyk H: Hyperplastic polyposis and the risk of colorectal cancer. Dis Colon Rectum 47 (12): 2101-4, 2004. [PUBMED Abstract]
- Koide N, Saito Y, Fujii T, et al.: A case of hyperplastic polyposis of the colon with adenocarcinomas in hyperplastic polyps after long-term follow-up. Endoscopy 34 (6): 499-502, 2002. [PUBMED Abstract]
- Jeevaratnam P, Cottier DS, Browett PJ, et al.: Familial giant hyperplastic polyposis predisposing to colorectal cancer: a new hereditary bowel cancer syndrome. J Pathol 179 (1): 20-5, 1996. [PUBMED Abstract]
- Bengoechea O, Martínez-Peñuela JM, Larrínaga B, et al.: Hyperplastic polyposis of the colorectum and adenocarcinoma in a 24-year-old man. Am J Surg Pathol 11 (4): 323-7, 1987. [PUBMED Abstract]
- McCann BG: A case of metaplastic polyposis of the colon associated with focal adenomatous change and metachronous adenocarcinomas. Histopathology 13 (6): 700-2, 1988. [PUBMED Abstract]
- Muller C, Yamada A, Ikegami S, et al.: Risk of Colorectal Cancer in Serrated Polyposis Syndrome: A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol 20 (3): 622-630.e7, 2022. [PUBMED Abstract]
- Kanth P, Yu Z, Keener MB, et al.: Cancer Risk in Patients With and Relatives of Serrated Polyposis Syndrome and Sporadic Sessile Serrated Lesions. Am J Gastroenterol 117 (2): 336-342, 2022. [PUBMED Abstract]
- Biller LH, Ukaegbu C, Dhingra TG, et al.: A Multi-Institutional Cohort of Therapy-Associated Polyposis in Childhood and Young Adulthood Cancer Survivors. Cancer Prev Res (Phila) 13 (3): 291-298, 2020. [PUBMED Abstract]
- Anthony E, Reece JC, Milanzi E, et al.: Body Mass Index, sex, non-steroidal anti-inflammatory drug medications, smoking and alcohol are differentially associated with World Health Organisation criteria and colorectal cancer risk in people with Serrated Polyposis Syndrome: an Australian case-control study. BMC Gastroenterol 22 (1): 489, 2022. [PUBMED Abstract]
- Hurlstone DP, Cross SS, Slater R, et al.: Detecting diminutive colorectal lesions at colonoscopy: a randomised controlled trial of pan-colonic versus targeted chromoscopy. Gut 53 (3): 376-80, 2004. [PUBMED Abstract]
- Saitoh Y, Waxman I, West AB, et al.: Prevalence and distinctive biologic features of flat colorectal adenomas in a North American population. Gastroenterology 120 (7): 1657-65, 2001. [PUBMED Abstract]
- Hurlstone DP, Cross SS, Adam I, et al.: Endoscopic morphological anticipation of submucosal invasion in flat and depressed colorectal lesions: clinical implications and subtype analysis of the kudo type V pit pattern using high-magnification-chromoscopic colonoscopy. Colorectal Dis 6 (5): 369-75, 2004. [PUBMED Abstract]
- Dacosta RS, Wilson BC, Marcon NE: New optical technologies for earlier endoscopic diagnosis of premalignant gastrointestinal lesions. J Gastroenterol Hepatol 17 (Suppl): S85-104, 2002. [PUBMED Abstract]
- Rembacken BJ, Fujii T, Cairns A, et al.: Flat and depressed colonic neoplasms: a prospective study of 1000 colonoscopies in the UK. Lancet 355 (9211): 1211-4, 2000. [PUBMED Abstract]
- Tsuda S, Veress B, Tóth E, et al.: Flat and depressed colorectal tumours in a southern Swedish population: a prospective chromoendoscopic and histopathological study. Gut 51 (4): 550-5, 2002. [PUBMED Abstract]
- Rex DK, Helbig CC: High yields of small and flat adenomas with high-definition colonoscopes using either white light or narrow band imaging. Gastroenterology 133 (1): 42-7, 2007. [PUBMED Abstract]
- Soetikno RM, Kaltenbach T, Rouse RV, et al.: Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA 299 (9): 1027-35, 2008. [PUBMED Abstract]
- Stoffel EM, Turgeon DK, Stockwell DH, et al.: Chromoendoscopy detects more adenomas than colonoscopy using intensive inspection without dye spraying. Cancer Prev Res (Phila) 1 (7): 507-13, 2008. [PUBMED Abstract]
- Le Rhun M, Coron E, Parlier D, et al.: High resolution colonoscopy with chromoscopy versus standard colonoscopy for the detection of colonic neoplasia: a randomized study. Clin Gastroenterol Hepatol 4 (3): 349-54, 2006. [PUBMED Abstract]
- Brooker JC, Saunders BP, Shah SG, et al.: Total colonic dye-spray increases the detection of diminutive adenomas during routine colonoscopy: a randomized controlled trial. Gastrointest Endosc 56 (3): 333-8, 2002. [PUBMED Abstract]
- Stoffel EM, Turgeon DK, Stockwell DH, et al.: Missed adenomas during colonoscopic surveillance in individuals with Lynch Syndrome (hereditary nonpolyposis colorectal cancer). Cancer Prev Res (Phila) 1 (6): 470-5, 2008. [PUBMED Abstract]
- Hüneburg R, Lammert F, Rabe C, et al.: Chromocolonoscopy detects more adenomas than white light colonoscopy or narrow band imaging colonoscopy in hereditary nonpolyposis colorectal cancer screening. Endoscopy 41 (4): 316-22, 2009. [PUBMED Abstract]
- Wallace MH, Frayling IM, Clark SK, et al.: Attenuated adenomatous polyposis coli: the role of ascertainment bias through failure to dye-spray at colonoscopy. Dis Colon Rectum 42 (8): 1078-80, 1999. [PUBMED Abstract]
- Dekker E, Boparai KS, Poley JW, et al.: High resolution endoscopy and the additional value of chromoendoscopy in the evaluation of duodenal adenomatosis in patients with familial adenomatous polyposis. Endoscopy 41 (8): 666-9, 2009. [PUBMED Abstract]
- Sakamoto H, Yamamoto H, Hayashi Y, et al.: Nonsurgical management of small-bowel polyps in Peutz-Jeghers syndrome with extensive polypectomy by using double-balloon endoscopy. Gastrointest Endosc 74 (2): 328-33, 2011. [PUBMED Abstract]
Psychosocial Issues in Hereditary Colon Cancer Syndromes
Introduction
Psychosocial research in cancer genetic counseling and testing focuses on the interest in testing among populations at varying levels of disease risk, psychological outcomes, interpersonal and familial effects, and cultural and community reactions. This research also identifies behavioral factors that encourage or impede surveillance and other health behaviors. Data resulting from psychosocial research can guide clinician interactions with patients and may include the following:
- Decision-making about risk-reduction interventions, risk assessment, and genetic testing.
- Evaluation of psychosocial interventions to reduce distress and/or other negative sequelae related to risk notification of genetic testing.
- Resolution of ethical concerns.
This section of the summary will focus on psychosocial aspects of genetic counseling and testing for Lynch syndrome, familial adenomatous polyposis (FAP), and Peutz-Jeghers syndrome (PJS), including issues surrounding medical screening, risk-reducing surgery, and chemoprevention for these syndromes.
Psychosocial Issues in Lynch Syndrome
Participation in genetic counseling and testing for Lynch syndrome
Early research on genetic counseling/testing uptake
Early studies that evaluated the uptake of genetic counseling and testing focused on selected, high-risk research populations, including colorectal cancer (CRC) patients and unaffected family members identified at high risk of CRC largely based on family history. The participants were recruited mainly from clinical settings and familial colon cancer registries. Most studies recruited index cancer cases, typically CRCs, specifically to offer genetic counseling and germline testing for mismatch repair (MMR) variants; these were frequently offered as free services.[1-9] Counseling and testing were similarly offered to relatives of index cases with pathogenic variants. A review that summarized these early studies reported a wide range of testing uptake rates, from 14% to 75%, and included uptake among both index cases and at-risk relatives who were offered testing.[10] The review indicated that the primary reasons for undergoing genetic testing included a desire to learn about children’s risk and to learn about early detection and screening needs, as well as a reduction in uncertainty. Reasons for declining testing included cost, insurance discrimination concerns, potential adverse emotional effects for oneself or one’s family, low anticipated benefit, and lack of time.
Uptake of genetic counseling and germline testing following universal tumor screening for microsatellite instability (MSI) and/or immunohistochemistry (IHC)
While these early studies of genetic testing uptake offered preliminary insight regarding why individuals may or may not be motivated to have testing, the process for offering genetic counseling and testing differed from what has evolved into current clinical practice. Clinical practice relies less solely on family history to identify individuals who may benefit from testing, and instead utilizes universal molecular diagnostic testing of CRC and endometrial cancer tumors in newly diagnosed patients using MSI and/or IHC as an initial screen for Lynch syndrome. (Refer to the Universal tumor testing to screen for Lynch syndrome section of this summary for more information.)
While universal MSI/IHC screening is increasingly being adopted to identify newly diagnosed patients who may have a germline variant, an important implication is that not all individuals who may be appropriate for germline testing follow through with recommended genetic counseling and testing services. Two reports from a single institution found that 20% and 13% of CRC and endometrial cancer index cases, respectively, with abnormal IHC results followed through with germline variant testing for Lynch syndrome.[11,12] These studies did not solicit reasons for follow through with genetic counseling and testing. However, it has been suggested that higher levels of patient completion of genetic testing after abnormal MSI/IHC results may be associated with having genetic counselors involved in this process to disclose screen-positive results, provide counseling after MSI/IHC testing, or facilitate referrals.[13]
In a study of 145 patients with CRC in the Kaiser Permanente Northwest health care system who were surveyed before receiving their MSI results, most patients had a positive attitude toward MSI/IHC screening.[14] The majority (84.8%) endorsed six or more benefits of MSI/IHC screening; however, 89.4% also endorsed fewer than four potential barriers, primarily the cost of additional testing and surveillance. Patients with stronger family histories of cancer were more likely to cite fewer barriers of MSI/IHC screening. Patients also experienced minimal distress associated with the screening, with 77.2% of participants having a score of zero (indicating no distress).
Education regarding family history and cancer risk and encouragement to have testing from health care providers may facilitate uptake of genetic counseling and testing. A small (n = 19) qualitative study of newly diagnosed patients with CRC who met high-risk criteria for referral to cancer genetics risk assessment and counseling identified potential reasons why patients may not seek counseling as recommended. These reasons included incomplete knowledge of family cancer history and not realizing the relevance of family history to their personal cancer diagnosis; lack of a specific, direct physician’s recommendation for counseling; and viewing counseling as a lower priority than coping with the immediate demands of a new cancer diagnosis.[15] In a follow-up survey of 91 individuals in a randomized trial to promote colonoscopy screening in those at risk for Lynch syndrome, only 24% reported ever having discussed genetic testing with their physicians, and the most common barrier to undergoing testing was lack of advice to do so by a health care provider.[16]
Uptake of cascade screening by at-risk relatives
There is increasing adoption of universal screening of newly diagnosed tumors for Lynch syndrome in clinical practice. However, the clinical benefit and cost-effectiveness of this process have been attributed to uptake of cascade screening, or predictive testing among at-risk relatives of index cancer cases who are found to have a pathogenic germline variant. A systematic review evaluated the frequency and predictors of genetic testing uptake by first-degree relatives (FDRs) of index cases with Lynch syndrome.[17] Among four studies that were included in the review and reported uptake rates among FDRs, results showed that 34% to 52% of FDRs had undergone testing. Factors associated with testing uptake in relatives included age (<50 y), female sex, parenthood, employment status, level of education, participation in medical research, psychological factors (lack of depressive symptoms), and the number of relatives affected with cancer.
A large retrospective study of genetic testing uptake across three generations of Finnish families enrolled in a Lynch syndrome registry also found an incomplete uptake of predictive testing among at-risk relatives of individuals with pathogenic variants, and a decreasing uptake rate by generation.[18] Among 1,184 probands with a Lynch syndrome variant, 67%, 43%, and 24% of at-risk adult first-, second-, and third-generation relatives, respectively, had predictive testing. Among 539 first-generation Lynch syndrome variant carriers, 62% of their at-risk adult children underwent testing. In multivariate analysis, older age, family-specific variant (MLH1 and MSH2 vs. MSH6), being an only child or having a sibling with a pathogenic variant, and having a parent who adhered to colonoscopy surveillance were associated with predictive testing uptake. This study suggested that family-level factors such as predictive testing and screening behavior may influence predictive testing among at-risk relatives of individuals with Lynch syndrome–associated variants.
Published reports of interventions to increase uptake of cascade screening in Lynch syndrome families are limited. An Australian paper compared two approaches for informing at-risk relatives about pathogenic variants for hereditary cancers, including Lynch syndrome.[19] In this study, index cases from 33 kindreds who had undergone genetic testing provided consent for their clinicians to send detailed letters to at-risk relatives advising them about the identification of an inherited cancer predisposition in the family. Letters also included a recommendation to discuss the information with a physician or genetics specialist, and they provided information about what a genetics evaluation comprised. Within the first 2 years of follow-up, 40% of first- and second-degree relatives (SDRs) had predictive genetic testing, were determined to be presumed noncarriers, or had undergone evaluation but declined genetic testing. The authors compared these findings with a cohort of 41 kindreds seen prior to the initiation of the clinician-generated letters, of whom variant-positive index cases had only been asked to advise relatives that genetic testing was available. In the earlier cohort, 23% of at-risk relatives had sought services to clarify their genetic risk status, which was significantly fewer compared with the group receiving clinician-generated letters (P = .001). Receipt of the letters did not generate concerns about a breach of privacy or autonomy.
Refer to the Ethical, Legal, and Social Implications section in the PDQ summary on Cancer Genetics Risk Assessment and Counseling for information about ethical concerns, including duty to warn.
Psychological impact of participating in genetic counseling and testing for Lynch syndrome
Studies have examined the psychological status of individuals before, during, and after genetic counseling and testing for Lynch syndrome. Some studies have included only persons with no personal history of any Lynch syndrome–associated cancers,[20-23] and others have included both CRC patients and cancer-unaffected persons who are at risk of having a Lynch syndrome pathogenic variant.[24-28] Cross-sectional evaluations of the psychosocial characteristics of individuals undergoing Lynch syndrome genetic counseling and testing have indicated that mean pretest scores of psychological functioning for most participants are within normal limits,[24-26] although one study comparing affected and unaffected individuals showed that affected individuals had greater distress and worry associated with Lynch syndrome.[29]
Several longitudinal studies have evaluated psychological outcomes before genetic counseling and testing for Lynch syndrome and at multiple time periods in the year after disclosure of test results. One study examined changes in anxiety based on personal cancer history, gender, and age (younger than 50 y vs. older than 50 y) before and 2 weeks after a pretest genetic-counseling session. Affected and unaffected female participants in both age groups and affected men older than 50 years showed significant decreases in anxiety over time. Unaffected men younger than 50 years maintained low levels of anxiety; however, affected men younger than 50 years showed no reductions in the anxiety levels reported at the time of pretest counseling.[30] A study that evaluated psychological distress 8 weeks postcounseling (before disclosure of test results) among both affected and unaffected individuals found a significant reduction in general anxiety, cancer worry, and distress.[29] In general, findings from studies within the time period immediately after disclosure of pathogenic variant status (e.g., 2 weeks to 1 month) suggested that carriers of mismatch repair (MMR) pathogenic variants may experience increased general distress,[22,27] cancer-specific distress,[20,21] or cancer worries [27] relative to their pretest measurements. Carriers often experienced significantly higher distress after disclosure of test results than do individuals who do not carry a pathogenic variant previously identified in the family (noncarrier).[20-22,27] However, in most cases, carriers’ distress levels subsided during the course of the year after disclosure [22,27] and did not differ from pretest distress levels at 1 year postdisclosure.[20,21] Findings from these studies also indicated that noncarriers experienced a reduction or no change in distress up to 1 year after results disclosure.[20-22,27] A study that included unaffected individuals and CRC patients found that distress levels among patients did not differ between carriers and individuals who received results that were uninformative or showed a variant of unknown significance at any point up to 1 year posttest and were similar compared with pretest distress levels.[28]
A limited number of studies have examined longer-term psychosocial outcomes after Lynch syndrome genetic counseling and testing.[20,31,32] Longitudinal studies that evaluated psychological distress before and after genetic testing found that long-term distress levels (measured at 3 or 7 years posttesting) among carriers and noncarriers of pathogenic variants were similar to distress levels at baseline.[20,32] with one exception: noncarriers’ cancer-specific distress scores in one study [20] showed a sustained decrease posttesting and were significantly lower than their baseline scores and with carriers’ scores at 1 year posttesting, with a similar trend observed at 3 years posttesting. In one study, carriers were more likely to be worried about CRC risk at 7 years posttesting; however, noncarriers who reported worry about CRC (i.e., “worried to some extent” or “very worried”) were more likely to doubt the validity of their test result than were noncarriers who reported no worry.[32] When asked about their satisfaction with the decision to have testing, the majority of carriers and noncarriers were extremely satisfied up to 7 years posttesting and indicated they would be willing to undergo testing again.[32]
Findings from some studies suggested that there may be subgroups of individuals at higher risk of psychological distress after disclosure of test results, including those who present with relatively higher scores on measures of general or cancer-specific distress before undergoing testing.[24-28,33] A study of CRC patients who had donated blood for Lynch syndrome testing found that higher levels of depressive symptoms and/or anxiety were found among women, younger individuals, non-White individuals, and those with less formal education and fewer and less satisfactory sources of social support.[24] A subgroup of individuals who showed higher levels of psychological distress and lower quality of life and social support were identified from the same population; in addition, this subgroup was more likely to worry about finding out that they were carriers of Lynch syndrome pathogenic variants and being able to cope with learning their test results.[25] In a follow-up report that evaluated psychological outcomes after the disclosure of test results among CRC patients and relatives at risk of having a Lynch syndrome pathogenic variant, a subgroup with the same psychosocial characteristics experienced higher levels of general distress and distress specific to the experience of having genetic testing within the year after disclosure, regardless of variant status. Non-White individuals and those with lower education had higher levels of depression and anxiety scores at all times compared with White individuals and those with higher education, respectively.[27] Other studies have also found that a prior history of major or minor depression, higher pretest levels of cancer-specific distress, having a greater number of cancer-affected FDRs, greater grief reactions, and greater emotional illness–related representations predicted higher levels of distress from 1 to 6 months after disclosure of test results.[28,33] While further research is needed in this area, case studies indicate that it is important to identify individuals who may be at risk of experiencing psychiatric distress and to provide psychological support and follow-up throughout the genetic counseling and genetic testing process.[34]
Studies also have examined the effect of Lynch syndrome genetic counseling and testing on cancer risk comprehension. One study reported that nearly all carriers and noncarriers of pathogenic variants could accurately recall the test result 1 year after disclosure. More noncarriers than carriers correctly identified their risk of developing CRC at both 1 month and 1 year after result disclosure. Carriers of pathogenic variants who incorrectly identified their CRC risk were more likely to have had lower levels of pretest subjective risk perception compared with those who correctly identified their level of risk.[22] Another study reported that accuracy of estimating colorectal and endometrial cancer risk improved after disclosure of variant status in carriers and noncarriers.[23]
Psychosocial aspects of screening and risk reduction interventions for Lynch syndrome
Screening
Colorectal screening for Lynch syndrome
Benefits of genetic counseling and testing for Lynch syndrome include the opportunity for individuals to learn about options for the early detection and prevention of cancer, including screening and risk-reducing surgery. Studies suggest that many individuals at risk of Lynch syndrome may have had some CRC screening before genetic counseling and testing, but most are not likely to adhere to Lynch syndrome screening recommendations. Among individuals aged 18 years or older who did not have a personal history of CRC and who participated in U.S.-based research protocols offering genetic counseling and testing for Lynch syndrome, between 52% and 62% reported ever having had a colonoscopy before genetic testing.[1,3,35,36] Among cancer-unaffected individuals who participated in similar research in Belgium and Australia, 51% and 68%, respectively, had ever had a colonoscopy before study entry.[23,37] Factors associated with ever having a colonoscopy before genetic testing included higher income and older age,[35] higher perceived risk of developing CRC,[37] higher education level, and being informed of increased risk of CRC.[36]
In a study of cancer-affected and cancer-unaffected individuals who fulfilled clinical criteria for Lynch syndrome, 92% reported having had a colonoscopy and/or flexible sigmoidoscopy at least once before genetic testing.[38] Another study of unaffected individuals presenting for genetic risk assessment and possible consideration of Lynch syndrome, FAP, or APC I1307K genetic testing reported that 77% had undergone at least one screening exam (either colonoscopy, flexible sigmoidoscopy, or barium enema).
Three studies determined whether cancer-unaffected individuals adhered to Lynch syndrome colonoscopy screening recommendations before genetic testing, and reported adherence rates of 10%,[23] 28%,[36] and 47%.[38]
Several longitudinal studies examined the use of screening colonoscopy by cancer-unaffected individuals after undergoing testing for a known Lynch syndrome pathogenic variant.[23,35-37] These studies compared colonoscopy use before Lynch syndrome genetic testing with colonoscopy use within 1 year after disclosure of test results. One study reported that carriers of Lynch syndrome pathogenic variants were more likely to have a colonoscopy than were noncarriers and those who declined testing (73% vs. 16% vs. 22%) and that colonoscopy use increased among carriers (36% vs. 73%) in the year after disclosure of results.[36] Two other studies reported that carriers’ colonoscopy rates at 1 year after disclosure of results (71% and 53%) were not significantly different from rates before testing,[35,37] although noncarriers’ colonoscopy rates decreased in the same time period. Factors associated with colonoscopy use at 1 year after disclosure of results included carrying a Lynch syndrome–predisposing pathogenic variant,[35-37] older age,[35] and greater perceived control over CRC. These findings suggest that colonoscopy rates increase or are maintained among carriers of pathogenic variants within the year after disclosure of results and that rates decrease among noncarriers. Data from a longitudinal study including 134 carriers of MMR pathogenic variants with and without a prior Lynch syndrome–related cancer diagnosis found that those who did not undergo colonoscopy for surveillance within 6 months after receiving genetic test results were six times more likely to report clinically significant depressive symptoms as measured by the Center for Epidemiological Studies-Depression (CES-D) scale (odds ratio [OR], 6.06; 95% confidence interval [CI], 2.09–17.59). Higher levels of CRC worry measured before genetic testing also were associated with clinically significant depressive symptoms (OR, 1.53; 95% CI, 1.19–1.97).[39]
Two studies examined the level of adherence to published screening guidelines after Lynch syndrome genetic testing, based on variant status. One study reported a colonoscopy adherence rate of 100% among carriers of pathogenic variants.[23] Another study found that 35% of carriers and 13% of noncarriers did not adhere to published guidelines for appropriate CRC screening;[35] in both groups, about one-half screened more frequently than published guidelines recommend, and one-half screened less frequently.
The longitudinal studies described above examined colorectal screening behavior within a relatively short period of time (1 year) after receiving genetic test results, and less is known about longer-term use of screening behaviors. A longitudinal study (N = 73) that examined psychological and behavioral outcomes among cancer-unaffected individuals at 3 years after disclosure of genetic test results found that all carriers (n = 19) had undergone at least one colonoscopy between 1 and 3 years postdisclosure.[20] A longitudinal study of similar outcomes up to 7 years posttesting also found that all carriers had undergone colonoscopy; most (83%) underwent the procedure every 3 years or more frequently as recommended, and 11% reported longer screening intervals.[32] In this study, those who reported longer screening intervals than recommended also were more likely to report a fear of dying soon. Also, 16% of noncarriers reported undergoing colonoscopy within the 7 years posttesting; those who indicated doubts about the validity of their test result were more likely to have had a colonoscopy.[32] Ninety-four percent of carriers in one study stated an intention to have annual or biannual colonoscopy in the future; among noncarriers, 64% did not intend to have colonoscopy in the future or were unsure, and 33% intended to have colonoscopy at 5- to 6-year intervals or less frequently.[23] A cross-sectional study conducted in the Netherlands examined the use of flexible sigmoidoscopy or colonoscopy among individuals with CRC, endometrial cancer, or a clinical or genetic diagnosis of Lynch syndrome during a time that ranged from 2 years to 18 years after risk assessment and counseling.[40] Eighty-six percent of carriers of Lynch syndrome pathogenic variants, 68% of those who did not test or who had an uninformative Lynch syndrome genetic test result, and 73% of those with a clinical Lynch syndrome diagnosis were considered adherent with screening recommendations, based on data obtained from medical records. Participants also answered questions regarding screening adherence, and 16% of the overall sample reported that they had undergone screening less frequently than recommended. For the overall sample, greater perceived barriers to screening were associated with screening nonadherence as determined through medical record review, and embarrassment with screening procedures was associated with self-reported nonadherence. A second cross-sectional study, also conducted in the Netherlands, surveyed cancer-unaffected carriers of Lynch syndrome variants (n = 42) regarding their colorectal screening behaviors after learning their pathogenic variant status (range, 6 mo–8.5 y). Thirty-one percent of respondents reported that they had undergone annual colonoscopy before Lynch syndrome genetic testing, and 88% reported that they had undergone colonoscopy since their genetic diagnosis (P < .001).[31]
Less is known about Lynch syndrome screening behaviors in individuals who may be at risk of having a germline pathogenic variant but who do not undergo genetic counseling and/or genetic testing to learn about their risk statuses. Among relatives of carriers of a Lynch syndrome germline pathogenic variant from the Australian Colorectal Cancer Family Registry, 26 who had not undergone genetic counseling and/or testing completed an interview to assess their perceived risk of developing CRC in the next 10 years and to self-report their colonoscopy status.[41] Their mean perceived risk was 30.5%, which exceeded the mean predicted risk of 4%, as calculated by MMRpro software.[42] Seventy-three percent (n = 19) reported undergoing a colonoscopy (one for diagnostic reasons); 35% had undergone colonoscopy within the past 2 years and had adhered to CRC screening recommendations. Perceived risk was slightly and positively correlated with years since an individual's last colonoscopy (Pearson's r, 0.49; range, 0.02–0.79) but otherwise was not associated with other screening or personal characteristics. The authors concluded that perceived CRC risk alone may not be a sufficient predictor of colonoscopy use in relatives of Lynch syndrome pathogenic variant carriers who have not undergone genetic counseling and/or testing.[41] Another study compared colonoscopy screening rates and predictors of CRC screening participation in two different populations: (1) people diagnosed with Lynch syndrome (n = 59), and (2) people at risk for Lynch syndrome who did not have genetic testing (n = 47). Individuals without genetic testing had lower colonoscopy participation rates than those with a Lynch syndrome pathogenic variant (47% vs. 73%, respectively). Perceived CRC risk and physician recommendation for CRC screening were important predictors of colonoscopy screening adherence, independent of an individual's genetic testing status.[43]
Gynecologic cancer screening for Lynch syndrome
Several small studies have examined the use of screening for endometrial and ovarian cancers associated with Lynch syndrome (refer to Table 16). There are several limitations to these studies, including small sample sizes, short follow-up, retrospective design, reliance on self-report as the data source, and some not including patients who had undergone Lynch syndrome genetic testing. Several studies have included individuals in the screening uptake analysis who do not meet the minimum age criteria for undergoing screening. Of the studies that assessed screening use after a negative test result for a known pathogenic variant in the family, only a few assessed indications for that screening, such as follow-up of a previously identified abnormality. Last, some studies have included patients in the uptake analysis who were actively undergoing treatment for another cancer, which could influence provider screening recommendations. Therefore, Table 16 is limited to studies with patients who had undergone Lynch syndrome genetic testing, larger sample sizes, longer follow-up, and analysis that included individuals of an appropriate screening age.
| Study Citation | Study Population | Uptake of Gynecologic Screening Before Genetic Counseling and Testing | Uptake of Gynecologic Screening After Receipt of Genetic Test Results | Length of Follow-up | Comments |
|---|---|---|---|---|---|
| EC = endometrial cancer; ES = endometrial sampling; RRH = risk-reducing total abdominal hysterectomy; RRSO = risk-reducing salpingo-oophorectomy; TVUS = transvaginal ultrasound. | |||||
| Noncarrier(s) = negative for known pathogenic variant in family. | |||||
| 1Prospective study design. | |||||
| 2Retrospective study design. | |||||
| aSelf-report as data source. | |||||
| Claes et al. (2005)1,a [23] | Carriers (n = 7) | Not reported | TVUS | 1 y | One noncarrier reported undergoing TVUS for a previous endometrial problem, while three noncarriers reported undergoing the procedure for preventive reasons |
| – Carriers 86% (6/7) | |||||
| Noncarriers (n = 16) | |||||
| – Noncarriers 27% (4/15) | |||||
| Collins et al. (2007)1,a [20] | Carriers (n = 13) | Not reported | TVUS | 3 y | Two of four carriers had an RRH/RRSO by the 3-year follow-up assessment |
| – Carriers 69% (9/13) | |||||
| – Noncarriers 6% (2/32) | |||||
| Noncarriers (n = 32) | ES | ||||
| – Carriers 54% (7/13) | |||||
| – Noncarriers 3% (1/32) | |||||
| Yurgelun et al. (2012): Cohort 12,a [44] | 77 at risk of Lynch syndrome–associated EC; 45 carriers; 19 no genetic testing but Lynch syndrome–associated family history | 75% (58/77) engaged in EC screening or EC risk-reduction intervention; 42 underwent annual TVUS and/or ES; 16 underwent RRH | Not reported | N/A | |
| Yurgelun et al. (2012): Cohort 21,a [44] | 40 women at clinical risk of Lynch syndrome | 65% (26/40) adhered to EC screening or risk reduction; 6 underwent RRH; 13 underwent annual ES and/or TVUS; 6 had not reached recommended screening age | Carriers: 100% (n = 16) adhered to EC screening or risk-reducing strategies; 4 underwent pretest RRH; 5 underwent RRH; 5 underwent EC screening (TVUS and/or ES); 2 had not reached recommended screening age | 1 y | |
| Carriers (n = 16) | |||||
| Noncarriers (n = 9); 14 indeterminate results; 1 variant of uncertain significance | Noncarriers: 11% (1/9) underwent EC screening; 11% (1/9) underwent RRH | ||||
Overall, these studies have included relatively small numbers of women and suggest that screening rates for Lynch syndrome–associated gynecologic cancers are low before genetic counseling and testing. However, after participation in genetic education and counseling and the receipt of Lynch syndrome pathogenic variant test results, uptake of gynecologic cancer screening in carriers generally increases, while noncarriers decrease use.
Risk-reducing surgery
There is no consensus regarding the use of risk-reducing colectomy for Lynch syndrome, and little is known about decision-making and psychological sequelae surrounding risk-reducing colectomy for Lynch syndrome.
Among individuals who received positive test results, a greater proportion indicated interest in having risk-reducing colectomy after disclosure of results than at baseline.[3] This study also indicated that consideration of risk-reducing surgery for Lynch syndrome may motivate participation in genetic testing. Before receiving results, 46% indicated that they were considering risk-reducing colectomy, and 69% of women were considering risk-reducing total abdominal hysterectomy (RRH) and risk reducing bilateral salpingo-oophorectomy (RRSO); however, this study did not assess whether individuals actually followed through with risk-reducing surgery after they received their test results. Before undergoing Lynch syndrome genetic counseling and testing, 5% of cancer-unaffected individuals at risk of an MMR variant in a longitudinal study reported that they would consider colectomy, and 5% of women indicated that they would have an RRH and an RRSO, if they were found to be pathogenic variant–positive. At 3 years after disclosure of results, no participants had undergone risk-reducing colectomy.[20,37] Two women who had undergone an RRH before genetic testing underwent RRSO within 1 year after testing,[37] however, no other female carriers of pathogenic variants in the study reported having either procedure at 3 years after test result disclosure.[20]
In a cross-sectional quality-of-life and functional outcome survey of Lynch syndrome patients with more extensive (subtotal colectomy) or less extensive (segmental resection or hemicolectomy) resections, global quality-of-life outcomes were comparable, although patients with greater extent of resection described more frequent bowel movements and related dysfunction.[45]
Family communication
Family communication about genetic testing for hereditary CRC susceptibility, and specifically about the results of such testing, is complex. It is generally accepted that communication about genetic risk information within families is largely the responsibility of family members themselves. A few studies have examined communication patterns in families who had been offered Lynch syndrome genetic counseling and testing. Studies have focused on whether individuals disclosed information about Lynch syndrome genetic testing to their family members, to whom they disclosed this information, and family-based characteristics or issues that might facilitate or inhibit such communication. These studies examined communication and disclosure processes in families after notification by health care professionals about a Lynch syndrome predisposition and have comprised relatively small samples.
Research findings indicate that individuals are generally willing to share information about the presence of a Lynch syndrome pathogenic variant within their families.[46-49] Motivations for sharing genetic risk information include a desire to increase family awareness about personal risk, health promotion options and predictive genetic testing, a desire for emotional support, and a perceived moral obligation and responsibility to help others in the family.[47-49] Findings across studies suggest that most study participants believed that Lynch syndrome genetic risk information is shared openly within families; however, such communication is more likely to occur with FDRs (e.g., siblings, children) than with more distant relatives.[46-49]
One Finnish study recruited parents aged 40 years or older and known to carry an MMR pathogenic variant to complete a questionnaire that investigated how parents shared knowledge of genetic risk with their adult and minor offspring. The study also identified challenges in the communication process.[50] Of 248 parents, 87% reported that they had disclosed results to their children. Reasons for nondisclosure were consistent with previous studies (young age of offspring, socially distant relationships, or feelings of difficulty in discussing the topic).[47,48,51] Nearly all parents had informed their adult offspring about their genetic risk and the possibility of genetic testing, but nearly one-third were unsure of how their offspring had used the information. Parents identified discussing their children’s cancer risk as the most difficult aspect of the communication process. Of the 191 firstborn children informed, 69% had undergone genetic testing. One-third of the parents suggested that health professionals should be involved in disclosure of the information and that a family appointment at the genetics clinic should be made at the time of disclosure.
In regard to informing second- and third-degree relatives, individuals may favor a cascade approach; for example, it is assumed that once a relative is given information about the family’s risk of Lynch syndrome, he or she would then be responsible for informing his or her FDRs.[46-48] This cascade approach to communication is distinctly preferred in regard to informing relatives’ offspring, particularly those of minor age, and the consensus suggests that it would be inappropriate to disclose such information to an SDR or third-degree relative without first proceeding through the family relational hierarchy.[46-48,51] In one study, individuals who had undergone testing and were found to carry a Lynch syndrome–predisposing pathogenic variant were more likely to inform at least one SDR or third-degree relative about their genetic test results than individuals who had received true negative or uninformative results.[49]
While communication about genetic risk is generally viewed as an open process, some communication barriers were reported across studies. Reasons for not informing a relative included lack of a close relationship and lack of contact with the individual; in fact, emotional, rather than relational, closeness seemed to be a more important determinant of the degree of risk communication. A desire to not worry relatives with information about test results and the perception that relatives would not understand the meaning of this information also have been cited as communication barriers.[49] Disclosure seemed less likely if at-risk individuals were considered too young to receive the information (i.e., children), if information about the hereditary cancer risk had previously created conflict in the family,[48] or if it was assumed that relatives would be uninterested in information about testing.[47] Prior existence of conflict seemed to inhibit discussions about hereditary cancer risk, particularly if such discussions involved disclosure of bad news.[48]
For most participants in these studies, the news that the pattern of cancers in their families was attributable to a Lynch syndrome–predisposing pathogenic variant did not come as a surprise,[46,47] as individuals had suspected a hereditary cause for the familial cancers or had prior family discussions about cancer. Identification of a Lynch syndrome–predisposing pathogenic variant in the family was considered a private matter but not necessarily a secret,[46] and many individuals had discussed the family’s pathogenic variant status with someone outside of the family. Knowledge about the detection of a Lynch syndrome–predisposing pathogenic variant in the family was not viewed as stigmatizing, though individuals expressed concern about the potential impact of this information on insurance discrimination.[46] Also, while there may be a willingness to disclose information about the presence of a pathogenic variant in the family, one study suggests a tendency to remain more private about the disclosure of individual results, distinguishing personal results from familial risk information.[51] In a few cases, individuals reported that their relatives expressed anger, shock, or other negative emotional reactions after receiving news about the family’s Lynch syndrome risk;[48] however, most indicated little to no difficulty in informing their relatives.[47] It was suggested that families who are more comfortable and open with cancer-related discussions may be more receptive and accepting of news about genetic risk.[48]
In some cases, probands reported feeling particularly obliged to inform family members about a hereditary cancer risk [48] and were often the strongest advocates for encouraging their family members to undergo genetic counseling and testing for the family pathogenic variant.[46] Some gender and family role differences also emerged in regard to the dissemination of hereditary cancer risk information. One study reported that female probands were more comfortable discussing genetic information than were male probands and that male probands showed a greater need for professional support during the family communication process.[47] Another study suggested that mothers may be particularly influential members of the family network in regard to communicating health risk information.[52] Pathogenic variant–negative individuals, individuals who chose not to be tested, and spouses of at-risk persons reported not feeling as personally involved with the risk communication process compared with probands and other at-risk persons who had undergone genetic testing.[46]
Various modes of communication (e.g., in-person, telephone, or written contact) may typically be used to disclose genetic risk information within families.[46-48] In one study, communication aids such as a genetic counseling summary letter or Lynch syndrome booklet were viewed as helpful adjuncts to the communication process but were not considered central or necessary to its success.[47] Studies have suggested that recommendations by health care providers to inform relatives about hereditary cancer risk may encourage communication about Lynch syndrome [48] and that support by health care professionals may be helpful in overcoming barriers to communicating such information to family members.[51]
Much of the literature to date on family communication has focused on disclosure of test results; however, other elements of family communication are currently being explored. One study evaluated the role of older family members in providing various types of support (e.g., instrumental, emotional, crisis help, and dependability when needed) among individuals with Lynch syndrome and their family members (206 respondents from 33 families).[7,53] Respondents completed interviews about their family social network (biological and non-biological relatives and others outside the family) and patterns of communication within their family. The median age of the respondents and the members of their family social network did not differ (age 43 y). The study found that 23% of the members of the family social network encouraged CRC screening (other types of support, such as social support, were reported much more frequently). Those who encouraged screening were older, female, and significant others or biological family members, rather than nonfamily members. Given that many of the members of the family social network did not live in the same household, the study points out the importance of extended family in the context of screening encouragement and support.
Psychosocial Issues in Familial Adenomatous Polyposis (FAP)
Participation in genetic counseling and testing for FAP
The uptake for genetic testing for FAP may be higher than testing for Lynch syndrome. A study of asymptomatic individuals in the United States at risk of FAP who were enrolled in a CRC registry and were offered genetic counseling found that 82% of adults and 95% of minors underwent genetic testing.[54] Uptake rates close to 100% have been reported in the United Kingdom.[55] A possible explanation for the greater uptake of APC genetic testing is that it may be more cost-effective than annual endoscopic screening [56] and can eliminate the burden of annual screening, which must often be initiated before puberty. The opportunity to eliminate worry about potential risk-reducing surgery is another possible benefit of genetic testing for FAP. The decision to have APC genetic testing may be viewed as a medical management decision;[57] the potential psychosocial factors that may influence the testing decision are not as well studied for FAP as for other hereditary cancer syndromes. The higher penetrance of APC pathogenic variants, earlier onset of disease, and the unambiguous phenotype also may influence the decision to undergo genetic testing for this condition, possibly because of a greater awareness of the disease and more experience with multiple family members being affected.
Genetic testing for FAP is presently offered to children with affected parents, often at the age of 10 to 12 years, when endoscopic screening is recommended. Because it is optimal to diagnose FAP before age 18 years to prevent CRC and because screening and possibly surgery are warranted at the time an individual is identified as a carrier of an APC pathogenic variant, genetic testing of minors is justified in this instance. (Refer to the Testing in children section in the PDQ summary on Cancer Genetics Risk Assessment and Counseling for a more detailed discussion regarding the ethical, psychosocial, and genetic counseling issues related to genetic testing in children.)
In a survey conducted in the Netherlands of members of families with FAP, one-third (34%) believed that it was most suitable to offer APC gene testing to children before age 12 years, whereas 38% preferred to offer testing to children between the ages of 12 and 16 years, when children would be better able to understand the DNA testing process. Only 4% felt that children should not undergo DNA testing at all.[58]
Results of qualitative interview data from 28 U.S. parents diagnosed with FAP showed that 61% favored genetic testing of APC variants in their at-risk children (aged 10–17 y); 71% believed that their children should receive their test results. The primary reasons why parents chose to test their children included early detection and management, reduction in parental anxiety and uncertainty, and help with decision making regarding surveillance. Reasons provided for not testing focused on discrimination concerns and cost.[59]
Clinical observations suggest that children who have family members affected with FAP are very aware of the possibility of risk-reducing surgery, and focus on the test result as the factor that determines the need for such surgery.[54] It is important to consider the timing of disclosure of genetic test results to children in regard to their age, developmental issues, and psychological concerns about FAP. Children who carry an APC pathogenic variant have expressed concern regarding how they will be perceived by peers and might benefit from assistance in formulating an explanation for others that preserves self-esteem.[54]
Psychological impact of participating in genetic counseling and testing for FAP
Studies evaluating psychological outcomes after genetic testing for FAP suggest that some individuals, particularly carriers of pathogenic variants, may be at risk of experiencing increased distress. In a cross-sectional study of adults who had previously undergone APC genetic testing, those who were carriers of pathogenic variants exhibited higher levels of state anxiety than noncarriers and were more likely to exhibit clinically significant anxiety levels.[60] Lower optimism and lower self-esteem were associated with higher anxiety in this study,[60] and FAP-related distress, perceived seriousness of FAP, and belief in the accuracy of genetic testing were associated with more state anxiety among carriers.[61] However, in an earlier study that compared adults who had undergone genetic testing for FAP, Huntington disease, and hereditary breast/ovarian cancer syndrome, FAP-specific distress was somewhat elevated within 1 week after disclosure of either positive or negative test results and was lower overall than the other syndromes.[57]
In a cross-sectional Australian study focusing on younger adults aged 18 to 35 years diagnosed with FAP (N = 88), participants most frequently reported the following FAP-related issues for which they perceived the need for moderate-to-high levels of support or assistance: anxiety regarding their children’s risk of developing FAP, fear about developing cancer, and uncertainty about the impact of FAP.[62] Seventy-five percent indicated that they would consider prenatal testing for FAP; 61% would consider PGT, and 61% would prefer that their children undergo genetic testing at birth or before age 10 years. A small proportion of respondents (16%) reported experiencing some FAP-related discrimination, primarily indicating that attending to their medical or self-care needs (e.g., time off work for screening, need for frequent toilet breaks, and physical limitations) may engender negative attitudes in colleagues and managers.
Another large cross-sectional study of FAP families conducted in the Netherlands included individuals aged 16 to 84 years who either had an FAP diagnosis, were at 50% risk of having an APC pathogenic variant, or were proven APC noncarriers.[63] Of those who had APC testing, 48% had done so at least 5 years or longer before this study. Of individuals with an FAP diagnosis, 76% had undergone preventive colectomy, and 78% of those were at least 5 years postsurgery. The study evaluated the prevalence of generalized psychological distress, distress related specifically to FAP, and cancer-related worries. Mean scores on the Mental Health Index-5, a subscale of the SF-36 that assessed generalized distress, were comparable to the general Dutch population. Twenty percent of respondents were classified as having moderate to high levels of FAP-specific distress as measured by the Impact of Event scale (IES), with 23% of those with an FAP diagnosis, 11% of those at risk of FAP, and 17% of noncarriers reporting scores in this range. Five percent reported scores on the IES that indicated severe and clinically relevant distress; of those, the majority (78%) had an FAP diagnosis. Overall, mean scores on the Cancer Worry Scale were comparable to those found in another study of families with Lynch syndrome. Individuals with an FAP diagnosis were more likely to report more frequent cancer worries, and the most commonly reported worries were the potential need for additional surgery (26%) and the likelihood that they (17%) or a family member (14%) will develop cancer. In multivariate analysis, factors associated with higher levels of FAP-specific distress included greater perceived risk of developing cancer, more frequent discussion about FAP with family or friends, and having no children. Factors associated with higher levels of cancer-specific worries included being female, poorer family functioning, greater actual and desired discussion about FAP with family or friends, greater perceived cancer risk, poorer general health perceptions, and having been a caregiver for a family member with cancer. The authors noted that most factors that were associated with higher levels of cancer- and FAP-specific distress or worry were psychosocial factors, rather than clinical or demographic factors.
Another cross-sectional study conducted in the Netherlands found that among FAP patients, 37% indicated that the disease had influenced their desire to have children (i.e., wanting fewer or no children). Thirty-three percent indicated that they would consider PND for FAP; 30% would consider PGT. Higher levels of guilt and more positive attitudes towards terminating pregnancy were associated with greater interest for both PND and PGT.[58] In a separate U.S. study, predictors of willingness to consider prenatal testing included having an affected child and experiencing an FDR’s death secondary to FAP.[64]
The psychological vulnerability of children undergoing testing is of particular concern in genetic testing for FAP. Research findings suggest that most children do not experience clinically significant psychological distress after APC testing. As in studies involving adults, however, subgroups may be vulnerable to increased distress and would benefit from continued psychological support. A study of children who had undergone genetic testing for FAP found that their mood and behavior remained in the normal range after genetic counseling and disclosure of test results. Aspects of the family situation, including illness in the mother or a sibling were associated with subclinical increases in depressive symptoms.[65] In a long-term follow-up study of 48 children undergoing testing for FAP, most children did not suffer psychological distress; however, a small proportion of children tested demonstrated clinically significant posttest distress.[66] Another study found that although APC pathogenic variant–positive children’s perceived risk of developing the disease increased after disclosure of results, anxiety and depression levels remain unchanged in the year after disclosure.[60] Pathogenic variant–negative children in this study experienced less anxiety and improved self-esteem over this same time period.
Psychosocial aspects of screening and risk reduction interventions for FAP
Screening
Colorectal screening for FAP
Less is known about psychological aspects of screening for FAP. One study of a small number of individuals (aged 17–53 y) with a family history of FAP who were offered participation in a genetic counseling and testing protocol found that among those who were asymptomatic, all reported undergoing at least one endoscopic surveillance before participation in the study.[38] Only 33% (two of six patients) reported continuing screening at the recommended interval. Of the affected individuals who had undergone colectomy, 92% (11 of 12 patients) were adherent to recommended colorectal surveillance. In a cross-sectional study of 150 individuals with a clinical or genetic diagnosis of classic FAP or attenuated FAP (AFAP) and at-risk relatives, 52% of those with FAP and 46% of relatives at risk of FAP, had undergone recommended endoscopic screening.[67] Among individuals who had or were at risk of AFAP, 58% and 33%, respectively, had undergone screening. Compared with individuals who had undergone screening within the recommended time interval, those who had not screened were less likely to recall provider recommendations for screening, more likely to lack health insurance or insurance reimbursement for screening, and more likely to believe that they are not at increased risk of CRC. Only 42% of the study population had ever undergone genetic counseling. A small percentage of participants (14%–19%) described screening as a “necessary evil,” indicating a dislike for the bowel preparation, or experienced pain and discomfort. Nineteen percent reported that these issues might pose barriers to undergoing future endoscopies. Nineteen percent reported that improved techniques and the use of anesthesia have improved tolerance for screening procedures.
Risk-reducing surgery
When individuals at risk of FAP develop multiple polyps, risk-reducing surgery in the form of subtotal colectomy or proctocolectomy is the only effective way to reduce the risk of CRC. Most persons with FAP can avoid a permanent ostomy and preserve the anus and/or rectum, allowing some degree of bowel continence. (Refer to the Interventions for FAP section of this summary for more information about surgical management procedures in FAP.) Evidence on the quality-of-life outcomes from these interventions continues to accumulate and is summarized in Table 17.
| Population | Length of Follow-up | Type of Procedure | Stool Frequency | Stool Continence | Body Image | Sexual Functioning | Comments |
|---|---|---|---|---|---|---|---|
| EORTC QLQ = European Organisation for Research and Treatment of Cancer Colorectal Quality of Life Questionnaire; IPAA = ileal pouch–anal anastomosis; IRA = ileorectal anastomosis; SD = standard deviation; SF-36 = Short Form (36) Health Survey. | |||||||
| aEORTC QLQ-C38 scores range from 0–100. Functional scales: 0 = lowest level of function and 100 = highest/healthy level of function. Symptom scales: 0 = lowest level of symptomatology and 100 = highest level of symptomatology. | |||||||
| bSF-36 scores range from 0–100, with 0 = lowest possible health status and 100 = best possible health status. | |||||||
| cWithin normal ranges for same age group. | |||||||
| 279 FAP-affected individuals (135 females and 144 males) after colectomy; controls included 1,771 individuals from the general Dutch population [68] | IRA mean: 12 y (SD, 7.5 y) | IRA: n = 161 | Not assessed | Not assessed | EORTC QLQ-CR38 a | EORTC QLQ-CR38 a | SF-36b scores (Dutch version) on all subscales were significantly lower than the scores in the general population (IRA: P < .001; IPAA: P < .001) |
| IRA: 87.5 (SD, 21.9) | IRA: 38.9 (SD, 26.6) | ||||||
| IPAA mean: 6.8 y (SD, 4.9 y) | IPAA: n = 118 | IPAA: 84.4 (SD, 22.7) | IPAA: 42.2 (SD, 26.3) | ||||
| 88 Australian individuals (63 females and 25 males) aged 18–35 y, including 57 after colectomy and 14 with FAP but no surgery [69] | Not reported | IRA: n = 33 | Not assessed | Not assessed | SF-36 b | SF-36 b | |
| IPAA: n = 21 | IRA: 89.9 (SD, 16.1) | IRA: 86.2 (SD, 21.6) | |||||
| Ileostomy: n = 1 | IPAA: 72.1 (SD, 23) | IPAA: 77.5 (SD, 26.2) | |||||
| Unknown surgery type: n = 2 | No surgery: 94.1 (SD, 9.4) | No surgery: 91 (SD, 19) | |||||
| 525 individuals (283 females and 242 males) including 296 after colectomy, 45 with FAP but no surgery, 50 at risk for FAP and no surgery, and 134 noncarriers [70] | Range: 0–1 y to >10 y | IRA: n = 136 | Not assessed | Not assessed | EORTC QLQ-CR38 a | EORTC QLQ-CR38 a | 41% of FAP patients reported employment disruptions: |
| After colectomy: 85.4 (SD, 20.5) | After colectomy: 42.2 (SD, 23.2) | Part or complete disability: n = 73 (59%) | |||||
| IPAA: n = 112 | FAP no surgery: 91.9 (SD, 16.1) | After colectomy: 42.2 (SD, 23.2) | Worked less: n = 30 (24%) | ||||
| Ileostomy: n = 42 | At risk: 94.0 (SD, 13.1) | At risk: 47.6 (SD, 23.7) | Worked more n = 5 (4%) | ||||
| Other: n = 6 | Noncarrier: 92.3 (SD, 13.1) | Noncarrier: 45.7 (SD, 21.2) | Worked more or less at different periods: n = 16 (13%) | ||||
| 209 Swedish FAP-affected individuals (116 females and 93 males) after colectomy aged 18–75 y [71] | Mean time since last surgery: 14 y (SD, 10; range, 1–50 y) | IRA: n = 71 | Not assessed | Day: 71% (n = 149) | Not assessed | Not assessed | The mean number of 21 abdominal symptoms assessed was 7 (SD, 4.61; range, 1–18). Women reported more symptoms than men, but there were no differences between genders regarding the degree the symptoms were troublesome. Higher symptom number was an independent predictor of poorer physical and mental health |
| IPAA: n = 82 | |||||||
| Ileostomy: n = 39 | Night: 61% (n = 128) | ||||||
| Continent ileostomy: n = 14 | |||||||
| Other: n = 3 | |||||||
| 28 individuals (10 females and 18 males) who underwent colectomy at age 14 y or younger [72] | 12 y (SD, 8.4; range, 1–37 y) | IRA: n = 7 | Day: | Day: | Rosenberg self-esteem score: 25.53/30c | Not assessed | 10/28 reported cancer-related worry post colectomy, with a trend that young age (<18 y) was associated with more cancer-related worry |
| IRA: 3.8 (SD, 1.5) | IRA: 71.4% (n = 7) | ||||||
| IPAA: 5.3 (SD, 2.4) | IPAA: 85.7% (n = 21) | ||||||
| IPAA: n = 21 | Night: | Night: | |||||
| IRA: 1.3 (SD, 0.6) | IRA: 50.0% (n = 7) | ||||||
| IPAA: 1.3 (SD, 0.5) | IPAA: 61.9% (n = 21) | ||||||
Studies of risk-reducing surgery for FAP have found that general measures of quality of life have been within normal range, and the majority reported no negative impact on their body image. However, these studies suggest that risk-reducing surgery for FAP may have negative quality-of-life effects for at least some proportion of those affected.
Chemoprevention
Chemoprevention trials are currently under way to evaluate the effectiveness of various therapies for individuals at risk of Lynch syndrome and FAP.[73,74] In a sample of individuals diagnosed with FAP who were invited to take part in a 5-year trial to evaluate the effects of vitamins and fiber on the development of adenomatous polyps, 55% agreed to participate.[75] Participants were more likely to be younger, to have been more recently diagnosed with FAP, and to live farther from the trial center, but did not differ from nonparticipants on any other psychosocial variables.
Reproductive Considerations in Individuals With Lynch Syndrome or FAP
Assisted reproductive technology (ART)
The possibility of transmitting a pathogenic variant to a child may pose a concern to families affected by hereditary CRC syndromes to the extent that some carriers may avoid childbearing. These concerns also may prompt individuals to consider using prenatal diagnosis (PND) methods to help reduce the risk of transmission. PND is an encompassing term used to refer to any medical procedure conducted to assess the presence of a genetic disorder in a fetus. Methods include amniocentesis and chorionic villous sampling.[76,77] Both procedures carry a small risk of miscarriage.[76,78] Moreover, discovering the fetus is a carrier of a cancer susceptibility variant may impose a difficult decision for couples regarding pregnancy continuation or termination and may require additional professional consultation and support.
An alternative to these tests is preimplantation genetic testing (PGT), a procedure used to test fertilized embryos for genetic disorders before uterine implantation.[79,80] Using the information obtained from the genetic testing, potential parents can decide whether or not to implant. PGT can be used to detect pathogenic variants in hereditary cancer predisposing genes, including APC.[58,64,81]
From the limited studies published to date, there appears to be interest in the use of ART for FAP, Lynch syndrome, and PJS.[58,64,82-84] However, actual uptake rates have not been reported.
| Study Population | Nc | Interest or Intention in ART | Comments |
|---|---|---|---|
| GT = genetic testing; PGT = preimplantation genetic testing; PND = prenatal diagnosis. | |||
| aStudies used a cross-sectional design and were conducted in the United States,[64] and in the Netherlands.[58,83]. | |||
| bParticipants were invited to complete questionnaires before clinical genetic testing for Lynch syndrome and at 3 months and 1 year after disclosure of genetic test results. | |||
| cIndicates number of participants older than 18 y, unless otherwise specified. | |||
| dTotal number of individuals with an APC pathogenic variant. Not all individuals answered or were eligible to answer each question. | |||
| eRepresents the number who indicated that they were considering having children in the future, out of a total of 130 individuals who answered a questionnaire before genetic testing.[82] | |||
| fTotal number of individuals with a Lynch syndrome pathogenic variant. Not all individuals answered or were eligible to answer each question. | |||
| FAP-affected individuals [64] | 20 | 95% (19/20) would consider prenatal GT for FAP; 90% (18/20) would consider PGT; 75% (15/20) would consider amniocentesis or chorionic villous sampling | |
| FAP-affected individuals [58] | 341 | 33% (16/64) would consider PND for FAP; 30% (76/256) would consider PGT; 15% (52/341) felt terminating pregnancy for FAP was acceptable | 24% and 25% of patients did not respond to questions about attitudes toward PND and PGT, respectively |
| Individuals with an APC pathogenic variant associated with FAP [84] | 65d | 25% (16/64) were aware of PGT; 78% (50/64) thought PGT should be offered; 55% (31/56) would consider PGT | |
| Individuals undergoing genetic testing for Lynch syndrome [82] | 48e | 21% 10/48) would consider PND and/or PGT; 19% (9/48) would consider only PND; 2% (1/48) would consider only PGT | At 1 year after disclosure of GT results, two of nine carriers reported that they were considering PGT for future pregnancy |
| Individuals with an identified Lynch syndrome pathogenic variant [84] | 43f | 19% (8/42) were aware of PGT; 69% (29/42) thought PGT should be offered; 41% (16/39) would consider PGT | |
| PJS-affected individualsa [83] | 52 | 15% (8/52) indicated that pregnancy termination was acceptable if PND identified a fetus with PJS; 52% (27/52) indicated PGT was acceptable for individuals with PJS | Ten (19%) individuals, nine of whom were female, reported that they had decided not to conceive a child because of PJS |
References
- Codori AM, Petersen GM, Miglioretti DL, et al.: Attitudes toward colon cancer gene testing: factors predicting test uptake. Cancer Epidemiol Biomarkers Prev 8 (4 Pt 2): 345-51, 1999. [PUBMED Abstract]
- Lerman C, Hughes C, Trock BJ, et al.: Genetic testing in families with hereditary nonpolyposis colon cancer. JAMA 281 (17): 1618-22, 1999. [PUBMED Abstract]
- Lynch HT, Lemon SJ, Karr B, et al.: Etiology, natural history, management and molecular genetics of hereditary nonpolyposis colorectal cancer (Lynch syndromes): genetic counseling implications. Cancer Epidemiol Biomarkers Prev 6 (12): 987-91, 1997. [PUBMED Abstract]
- Vernon SW, Gritz ER, Peterson SK, et al.: Intention to learn results of genetic testing for hereditary colon cancer. Cancer Epidemiol Biomarkers Prev 8 (4 Pt 2): 353-60, 1999. [PUBMED Abstract]
- Aktan-Collan K, Mecklin JP, Järvinen H, et al.: Predictive genetic testing for hereditary non-polyposis colorectal cancer: uptake and long-term satisfaction. Int J Cancer 89 (1): 44-50, 2000. [PUBMED Abstract]
- Loader S, Shields C, Levenkron JC, et al.: Patient vs. physician as the target of educational outreach about screening for an inherited susceptibility to colorectal cancer. Genet Test 6 (4): 281-90, 2002. [PUBMED Abstract]
- Hadley DW, Jenkins J, Dimond E, et al.: Genetic counseling and testing in families with hereditary nonpolyposis colorectal cancer. Arch Intern Med 163 (5): 573-82, 2003. [PUBMED Abstract]
- Keller M, Jost R, Kadmon M, et al.: Acceptance of and attitude toward genetic testing for hereditary nonpolyposis colorectal cancer: a comparison of participants and nonparticipants in genetic counseling. Dis Colon Rectum 47 (2): 153-62, 2004. [PUBMED Abstract]
- Johnson KA, Rosenblum-Vos L, Petersen GM, et al.: Response to genetic counseling and testing for the APC I1307K mutation. Am J Med Genet 91 (3): 207-11, 2000. [PUBMED Abstract]
- Bleiker EM, Esplen MJ, Meiser B, et al.: 100 years Lynch syndrome: what have we learned about psychosocial issues? Fam Cancer 12 (2): 325-39, 2013. [PUBMED Abstract]
- South CD, Yearsley M, Martin E, et al.: Immunohistochemistry staining for the mismatch repair proteins in the clinical care of patients with colorectal cancer. Genet Med 11 (11): 812-7, 2009. [PUBMED Abstract]
- Backes FJ, Leon ME, Ivanov I, et al.: Prospective evaluation of DNA mismatch repair protein expression in primary endometrial cancer. Gynecol Oncol 114 (3): 486-90, 2009. [PUBMED Abstract]
- Cragun D, DeBate RD, Vadaparampil ST, et al.: Comparing universal Lynch syndrome tumor-screening programs to evaluate associations between implementation strategies and patient follow-through. Genet Med 16 (10): 773-82, 2014. [PUBMED Abstract]
- Hunter JE, Zepp JM, Gilmore MJ, et al.: Universal tumor screening for Lynch syndrome: Assessment of the perspectives of patients with colorectal cancer regarding benefits and barriers. Cancer 121 (18): 3281-9, 2015. [PUBMED Abstract]
- Tomiak E, Samson A, Spector N, et al.: Reflex testing for Lynch syndrome: if we build it, will they come? Lessons learned from the uptake of clinical genetics services by individuals with newly diagnosed colorectal cancer (CRC). Fam Cancer 13 (1): 75-82, 2014. [PUBMED Abstract]
- Patel SG, Ahnen DJ, Kinney AY, et al.: Knowledge and Uptake of Genetic Counseling and Colonoscopic Screening Among Individuals at Increased Risk for Lynch Syndrome and their Endoscopists from the Family Health Promotion Project. Am J Gastroenterol 111 (2): 285-93, 2016. [PUBMED Abstract]
- Sharaf RN, Myer P, Stave CD, et al.: Uptake of genetic testing by relatives of lynch syndrome probands: a systematic review. Clin Gastroenterol Hepatol 11 (9): 1093-100, 2013. [PUBMED Abstract]
- Seppälä TT, Pylvänäinen K, Mecklin JP: Uptake of genetic testing by the children of Lynch syndrome variant carriers across three generations. Eur J Hum Genet 25 (11): 1237-1245, 2017. [PUBMED Abstract]
- Suthers GK, Armstrong J, McCormack J, et al.: Letting the family know: balancing ethics and effectiveness when notifying relatives about genetic testing for a familial disorder. J Med Genet 43 (8): 665-70, 2006. [PUBMED Abstract]
- Collins VR, Meiser B, Ukoumunne OC, et al.: The impact of predictive genetic testing for hereditary nonpolyposis colorectal cancer: three years after testing. Genet Med 9 (5): 290-7, 2007. [PUBMED Abstract]
- Meiser B, Collins V, Warren R, et al.: Psychological impact of genetic testing for hereditary non-polyposis colorectal cancer. Clin Genet 66 (6): 502-11, 2004. [PUBMED Abstract]
- Aktan-Collan K, Haukkala A, Mecklin JP, et al.: Psychological consequences of predictive genetic testing for hereditary non-polyposis colorectal cancer (HNPCC): a prospective follow-up study. Int J Cancer 93 (4): 608-11, 2001. [PUBMED Abstract]
- Claes E, Denayer L, Evers-Kiebooms G, et al.: Predictive testing for hereditary nonpolyposis colorectal cancer: subjective perception regarding colorectal and endometrial cancer, distress, and health-related behavior at one year post-test. Genet Test 9 (1): 54-65, 2005. [PUBMED Abstract]
- Vernon SW, Gritz ER, Peterson SK, et al.: Correlates of psychologic distress in colorectal cancer patients undergoing genetic testing for hereditary colon cancer. Health Psychol 16 (1): 73-86, 1997. [PUBMED Abstract]
- Gritz ER, Vernon SW, Peterson SK, et al.: Distress in the cancer patient and its association with genetic testing and counseling for hereditary non-polyposis colon cancer. Cancer Research, Therapy and Control 8(1-2): 35-49, 1999.
- Esplen MJ, Urquhart C, Butler K, et al.: The experience of loss and anticipation of distress in colorectal cancer patients undergoing genetic testing. J Psychosom Res 55 (5): 427-35, 2003. [PUBMED Abstract]
- Gritz ER, Peterson SK, Vernon SW, et al.: Psychological impact of genetic testing for hereditary nonpolyposis colorectal cancer. J Clin Oncol 23 (9): 1902-10, 2005. [PUBMED Abstract]
- Murakami Y, Okamura H, Sugano K, et al.: Psychologic distress after disclosure of genetic test results regarding hereditary nonpolyposis colorectal carcinoma. Cancer 101 (2): 395-403, 2004. [PUBMED Abstract]
- Keller M, Jost R, Haunstetter CM, et al.: Psychosocial outcome following genetic risk counselling for familial colorectal cancer. A comparison of affected patients and family members. Clin Genet 74 (5): 414-24, 2008. [PUBMED Abstract]
- Hasenbring MI, Kreddig N, Deges G, et al.: Psychological impact of genetic counseling for hereditary nonpolyposis colorectal cancer: the role of cancer history, gender, age, and psychological distress. Genet Test Mol Biomarkers 15 (4): 219-25, 2011. [PUBMED Abstract]
- Wagner A, van Kessel I, Kriege MG, et al.: Long term follow-up of HNPCC gene mutation carriers: compliance with screening and satisfaction with counseling and screening procedures. Fam Cancer 4 (4): 295-300, 2005. [PUBMED Abstract]
- Aktan-Collan K, Kääriäinen H, Järvinen H, et al.: Psychosocial consequences of predictive genetic testing for Lynch syndrome and associations to surveillance behaviour in a 7-year follow-up study. Fam Cancer 12 (4): 639-46, 2013. [PUBMED Abstract]
- van Oostrom I, Meijers-Heijboer H, Duivenvoorden HJ, et al.: Experience of parental cancer in childhood is a risk factor for psychological distress during genetic cancer susceptibility testing. Ann Oncol 17 (7): 1090-5, 2006. [PUBMED Abstract]
- Patenaude AF: Genetic Testing for Cancer: Psychological Approaches for Helping Patients and Families. American Psychological Association, 2005.
- Hadley DW, Jenkins JF, Dimond E, et al.: Colon cancer screening practices after genetic counseling and testing for hereditary nonpolyposis colorectal cancer. J Clin Oncol 22 (1): 39-44, 2004. [PUBMED Abstract]
- Halbert CH, Lynch H, Lynch J, et al.: Colon cancer screening practices following genetic testing for hereditary nonpolyposis colon cancer (HNPCC) mutations. Arch Intern Med 164 (17): 1881-7, 2004. [PUBMED Abstract]
- Collins V, Meiser B, Gaff C, et al.: Screening and preventive behaviors one year after predictive genetic testing for hereditary nonpolyposis colorectal carcinoma. Cancer 104 (2): 273-81, 2005. [PUBMED Abstract]
- Stoffel EM, Garber JE, Grover S, et al.: Cancer surveillance is often inadequate in people at high risk for colorectal cancer. J Med Genet 40 (5): e54, 2003. [PUBMED Abstract]
- Hadley DW, Ashida S, Jenkins JF, et al.: Colonoscopy use following mutation detection in Lynch syndrome: exploring a role for cancer screening in adaptation. Clin Genet 79 (4): 321-8, 2011. [PUBMED Abstract]
- Bleiker EM, Menko FH, Taal BG, et al.: Screening behavior of individuals at high risk for colorectal cancer. Gastroenterology 128 (2): 280-7, 2005. [PUBMED Abstract]
- Flander L, Speirs-Bridge A, Rutstein A, et al.: Perceived versus predicted risks of colorectal cancer and self-reported colonoscopies by members of mismatch repair gene mutation-carrying families who have declined genetic testing. J Genet Couns 23 (1): 79-88, 2014. [PUBMED Abstract]
- Chen S, Wang W, Lee S, et al.: Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 296 (12): 1479-87, 2006. [PUBMED Abstract]
- Hadley DW, Eliezer D, Addissie Y, et al.: Uptake and predictors of colonoscopy use in family members not participating in cascade genetic testing for Lynch syndrome. Sci Rep 10 (1): 15959, 2020. [PUBMED Abstract]
- Yurgelun MB, Mercado R, Rosenblatt M, et al.: Impact of genetic testing on endometrial cancer risk-reducing practices in women at risk for Lynch syndrome. Gynecol Oncol 127 (3): 544-51, 2012. [PUBMED Abstract]
- Haanstra JF, de Vos Tot Nederveen Cappel WH, Gopie JP, et al.: Quality of life after surgery for colon cancer in patients with Lynch syndrome: partial versus subtotal colectomy. Dis Colon Rectum 55 (6): 653-9, 2012. [PUBMED Abstract]
- Peterson SK, Watts BG, Koehly LM, et al.: How families communicate about HNPCC genetic testing: findings from a qualitative study. Am J Med Genet C Semin Med Genet 119 (1): 78-86, 2003. [PUBMED Abstract]
- Gaff CL, Collins V, Symes T, et al.: Facilitating family communication about predictive genetic testing: probands' perceptions. J Genet Couns 14 (2): 133-40, 2005. [PUBMED Abstract]
- Mesters I, Ausems M, Eichhorn S, et al.: Informing one's family about genetic testing for hereditary non-polyposis colorectal cancer (HNPCC): a retrospective exploratory study. Fam Cancer 4 (2): 163-7, 2005. [PUBMED Abstract]
- Stoffel EM, Ford B, Mercado RC, et al.: Sharing genetic test results in Lynch syndrome: communication with close and distant relatives. Clin Gastroenterol Hepatol 6 (3): 333-8, 2008. [PUBMED Abstract]
- Aktan-Collan KI, Kääriäinen HA, Kolttola EM, et al.: Sharing genetic risk with next generation: mutation-positive parents' communication with their offspring in Lynch Syndrome. Fam Cancer 10 (1): 43-50, 2011. [PUBMED Abstract]
- Pentz RD, Peterson SK, Watts B, et al.: Hereditary nonpolyposis colorectal cancer family members' perceptions about the duty to inform and health professionals' role in disseminating genetic information. Genet Test 9 (3): 261-8, 2005. [PUBMED Abstract]
- Koehly LM, Peterson SK, Watts BG, et al.: A social network analysis of communication about hereditary nonpolyposis colorectal cancer genetic testing and family functioning. Cancer Epidemiol Biomarkers Prev 12 (4): 304-13, 2003. [PUBMED Abstract]
- Ashida S, Hadley DW, Goergen AF, et al.: The importance of older family members in providing social resources and promoting cancer screening in families with a hereditary cancer syndrome. Gerontologist 51 (6): 833-42, 2011. [PUBMED Abstract]
- Petersen GM, Boyd PA: Gene tests and counseling for colorectal cancer risk: lessons from familial polyposis. J Natl Cancer Inst Monogr (17): 67-71, 1995. [PUBMED Abstract]
- Whitelaw S, Northover JM, Hodgson SV: Attitudes to predictive DNA testing in familial adenomatous polyposis. J Med Genet 33 (7): 540-3, 1996. [PUBMED Abstract]
- Bapat B, Noorani H, Cohen Z, et al.: Cost comparison of predictive genetic testing versus conventional clinical screening for familial adenomatous polyposis. Gut 44 (5): 698-703, 1999. [PUBMED Abstract]
- Dudok deWit AC, Duivenvoorden HJ, Passchier J, et al.: Course of distress experienced by persons at risk for an autosomal dominant inheritable disorder participating in a predictive testing program: an explorative study. Rotterdam/Leiden Genetics Workgroup. Psychosom Med 60 (5): 543-9, 1998 Sep-Oct. [PUBMED Abstract]
- Douma KF, Aaronson NK, Vasen HF, et al.: Attitudes toward genetic testing in childhood and reproductive decision-making for familial adenomatous polyposis. Eur J Hum Genet 18 (2): 186-93, 2010. [PUBMED Abstract]
- Levine FR, Coxworth JE, Stevenson DA, et al.: Parental attitudes, beliefs, and perceptions about genetic testing for FAP and colorectal cancer surveillance in minors. J Genet Couns 19 (3): 269-79, 2010. [PUBMED Abstract]
- Michie S, Bobrow M, Marteau TM: Predictive genetic testing in children and adults: a study of emotional impact. J Med Genet 38 (8): 519-26, 2001. [PUBMED Abstract]
- Michie S, Weinman J, Miller J, et al.: Predictive genetic testing: high risk expectations in the face of low risk information. J Behav Med 25 (1): 33-50, 2002. [PUBMED Abstract]
- Andrews L, Mireskandari S, Jessen J, et al.: Impact of familial adenomatous polyposis on young adults: attitudes toward genetic testing, support, and information needs. Genet Med 8 (11): 697-703, 2006. [PUBMED Abstract]
- Douma KF, Aaronson NK, Vasen HF, et al.: Psychological distress and use of psychosocial support in familial adenomatous polyposis. Psychooncology 19 (3): 289-98, 2010. [PUBMED Abstract]
- Kastrinos F, Stoffel EM, Balmaña J, et al.: Attitudes toward prenatal genetic testing in patients with familial adenomatous polyposis. Am J Gastroenterol 102 (6): 1284-90, 2007. [PUBMED Abstract]
- Codori AM, Petersen GM, Boyd PA, et al.: Genetic testing for cancer in children. Short-term psychological effect. Arch Pediatr Adolesc Med 150 (11): 1131-8, 1996. [PUBMED Abstract]
- Codori AM, Zawacki KL, Petersen GM, et al.: Genetic testing for hereditary colorectal cancer in children: long-term psychological effects. Am J Med Genet 116A (2): 117-28, 2003. [PUBMED Abstract]
- Kinney AY, Hicken B, Simonsen SE, et al.: Colorectal cancer surveillance behaviors among members of typical and attenuated FAP families. Am J Gastroenterol 102 (1): 153-62, 2007. [PUBMED Abstract]
- Van Duijvendijk P, Slors JF, Taat CW, et al.: Quality of life after total colectomy with ileorectal anastomosis or proctocolectomy and ileal pouch-anal anastomosis for familial adenomatous polyposis. Br J Surg 87 (5): 590-6, 2000. [PUBMED Abstract]
- Andrews L, Mireskandari S, Jessen J, et al.: Impact of familial adenomatous polyposis on young adults: quality of life outcomes. Dis Colon Rectum 50 (9): 1306-15, 2007. [PUBMED Abstract]
- Douma KF, Bleiker EM, Vasen HF, et al.: Quality of life and consequences for daily life of familial adenomatous polyposis (FAP) family members. Colorectal Dis 13 (6): 669-77, 2011. [PUBMED Abstract]
- Fritzell K, Eriksson LE, Björk J, et al.: Self-reported abdominal symptoms in relation to health status in adult patients with familial adenomatous polyposis. Dis Colon Rectum 54 (7): 863-9, 2011. [PUBMED Abstract]
- Durno CA, Wong J, Berk T, et al.: Quality of life and functional outcome for individuals who underwent very early colectomy for familial adenomatous polyposis. Dis Colon Rectum 55 (4): 436-43, 2012. [PUBMED Abstract]
- Hawk E, Lubet R, Limburg P: Chemoprevention in hereditary colorectal cancer syndromes. Cancer 86 (11 Suppl): 2551-63, 1999. [PUBMED Abstract]
- Celecoxib trials under Way J Natl Cancer Inst 92 (4): 299A-299, 2000. [PUBMED Abstract]
- Miller HH, Bauman LJ, Friedman DR, et al.: Psychosocial adjustment of familial polyposis patients and participation in a chemoprevention trial. Int J Psychiatry Med 16 (3): 211-30, 1986-87. [PUBMED Abstract]
- Cunniff C; American Academy of Pediatrics Committee on Genetics: Prenatal screening and diagnosis for pediatricians. Pediatrics 114 (3): 889-94, 2004. [PUBMED Abstract]
- Rappaport VJ: Prenatal diagnosis and genetic screening--integration into prenatal care. Obstet Gynecol Clin North Am 35 (3): 435-58, ix, 2008. [PUBMED Abstract]
- Eddleman KA, Malone FD, Sullivan L, et al.: Pregnancy loss rates after midtrimester amniocentesis. Obstet Gynecol 108 (5): 1067-72, 2006. [PUBMED Abstract]
- Baruch S, Kaufman D, Hudson KL: Genetic testing of embryos: practices and perspectives of US in vitro fertilization clinics. Fertil Steril 89 (5): 1053-8, 2008. [PUBMED Abstract]
- Ogilvie CM, Braude PR, Scriven PN: Preimplantation genetic diagnosis--an overview. J Histochem Cytochem 53 (3): 255-60, 2005. [PUBMED Abstract]
- Simpson JL, Carson SA, Cisneros P: Preimplantation genetic diagnosis (PGD) for heritable neoplasia. J Natl Cancer Inst Monogr (34): 87-90, 2005. [PUBMED Abstract]
- Dewanwala A, Chittenden A, Rosenblatt M, et al.: Attitudes toward childbearing and prenatal testing in individuals undergoing genetic testing for Lynch syndrome. Fam Cancer 10 (3): 549-56, 2011. [PUBMED Abstract]
- van Lier MG, Korsse SE, Mathus-Vliegen EM, et al.: Peutz-Jeghers syndrome and family planning: the attitude towards prenatal diagnosis and pre-implantation genetic diagnosis. Eur J Hum Genet 20 (2): 236-9, 2012. [PUBMED Abstract]
- Rich TA, Liu M, Etzel CJ, et al.: Comparison of attitudes regarding preimplantation genetic diagnosis among patients with hereditary cancer syndromes. Fam Cancer 13 (2): 291-9, 2014. [PUBMED Abstract]
Latest Updates to This Summary (03/21/2025)
The PDQ cancer information summaries are reviewed regularly and updated as new information becomes available. This section describes the latest changes made to this summary as of the date above.
Updated National Comprehensive Cancer Network (NCCN) as reference 56.
Revised text to state that if at-risk minors are not tested, high-quality colonoscopy is initiated between ages 10 to 15 years (cited NCCN [Genetic/Familial High-Risk Assessment: Colorectal, Endometrial, and Gastric] as reference 123).
Updated Table 7, Clinical Practice Guidelines for Colon Surveillance of Attenuated Familial Adenomatous Polyposis, with new NCCN recommendations.
Revised text to state that clinical management guidelines for MUTYH-associated polyposis (MAP) recommend screening with high-quality colonoscopy and polypectomy every 1 to 2 years if fewer than 20 adenomas are found.
Updated Table 8, Clinical Practice Guidelines for Colon Surveillance of MAP, with new NCCN recommendations.
Updated Table 13, Gene-Specific Practice Guidelines for Colon Surveillance of Lynch Syndrome, with new NCCN recommendations.
Revised text to state that transvaginal ultrasound is not recommended for endometrial cancer screening in premenopausal patients with Lynch syndrome.
Revised text to state that NCCN does not recommend routine ovarian cancer screening in individuals with Lynch syndrome. However, CA-125 and pelvic ultrasound are recommended for planning risk-reducing surgery.
Revised text to state that screening can be initiated before age 30 years if any of the following are present: a family history of cancers in the upper gastrointestinal (GI) system, incomplete or extensive gastric intestinal metaplasia, gastric/duodenal adenomas, or Barrett's esophagus with dysplasia. Other risk factors for gastric cancer include the following: male sex, older age, an MLH1 or MSH2 pathogenic variant, Asian ethnicity, residing in or emigrating from countries with a high background incidence of gastric cancer, and chronic autoimmune gastritis.
Added text about new NCCN recommendations for POLD1 and POLE carriers.
Added text about CRC and breast cancer screening recommendations for biallelic NTHL1 carriers.
Added text to state that consistent with prior reports, monoallelic NTHL1 carriers do not have significantly increased cancer risk. It is not recommended that these individuals undergo intensive cancer surveillance for GI, breast, or gynecologic cancers.
Added text about new NCCN recommendations for AXIN2 carriers.
Added text about new NCCN recommendations for GREM1 carriers.
Added text about new NCCN recommendations for CHEK2 carriers.
Added text about new NCCN recommendations for individuals with serrated polyposis syndrome.
This summary is written and maintained by the PDQ Cancer Genetics Editorial Board, which is editorially independent of NCI. The summary reflects an independent review of the literature and does not represent a policy statement of NCI or NIH. More information about summary policies and the role of the PDQ Editorial Boards in maintaining the PDQ summaries can be found on the About This PDQ Summary and PDQ® Cancer Information for Health Professionals pages.
About This PDQ Summary
Purpose of This Summary
This PDQ cancer information summary for health professionals provides comprehensive, peer-reviewed, evidence-based information about the genetics of colorectal cancer. It is intended as a resource to inform and assist clinicians in the care of their patients. It does not provide formal guidelines or recommendations for making health care decisions.
Reviewers and Updates
This summary is reviewed regularly and updated as necessary by the PDQ Cancer Genetics Editorial Board, which is editorially independent of the National Cancer Institute (NCI). The summary reflects an independent review of the literature and does not represent a policy statement of NCI or the National Institutes of Health (NIH).
Board members review recently published articles each month to determine whether an article should:
- be discussed at a meeting,
- be cited with text, or
- replace or update an existing article that is already cited.
Changes to the summaries are made through a consensus process in which Board members evaluate the strength of the evidence in the published articles and determine how the article should be included in the summary.
The lead reviewers for Genetics of Colorectal Cancer are:
- Melyssa D. Aronson, MS, CGC, CCGC (Sinai Health System)
- Kathleen A. Calzone, PhD, RN, AGN-BC, FAAN (National Cancer Institute)
- Zachariah Foda, MD, PhD (Johns Hopkins University School of Medicine)
- Katharine Germansky, MD (Beth Israel Deaconess Medical Center)
- David Liska, MD, MA (Cleveland Clinic)
- Gautam Mankaney, MD (Virginia Mason Franciscan Health)
- Suzanne C. O'Neill, PhD (Georgetown University)
- John M. Quillin, PhD, MPH, MS (Virginia Commonwealth University)
- Charite Ricker, MS, CGC (University of Southern California)
- Catharine Wang, PhD, MSc (Boston University School of Public Health)
- Matthew B. Yurgelun, MD (Dana-Farber Cancer Institute)
- Kevin Zbuk, MD, FRCPC (Margaret and Charles Juravinski Cancer Centre)
Any comments or questions about the summary content should be submitted to Cancer.gov through the NCI website's Email Us. Do not contact the individual Board Members with questions or comments about the summaries. Board members will not respond to individual inquiries.
Levels of Evidence
Some of the reference citations in this summary are accompanied by a level-of-evidence designation. These designations are intended to help readers assess the strength of the evidence supporting the use of specific interventions or approaches. The PDQ Cancer Genetics Editorial Board uses a formal evidence ranking system in developing its level-of-evidence designations.
Permission to Use This Summary
PDQ is a registered trademark. Although the content of PDQ documents can be used freely as text, it cannot be identified as an NCI PDQ cancer information summary unless it is presented in its entirety and is regularly updated. However, an author would be permitted to write a sentence such as “NCI’s PDQ cancer information summary about breast cancer prevention states the risks succinctly: [include excerpt from the summary].”
The preferred citation for this PDQ summary is:
PDQ® Cancer Genetics Editorial Board. PDQ Genetics of Colorectal Cancer. Bethesda, MD: National Cancer Institute. Updated <MM/DD/YYYY>. Available at: https://www.cancer.gov/types/colorectal/hp/colorectal-genetics-pdq. Accessed <MM/DD/YYYY>. [PMID: 26389505]
Images in this summary are used with permission of the author(s), artist, and/or publisher for use within the PDQ summaries only. Permission to use images outside the context of PDQ information must be obtained from the owner(s) and cannot be granted by the National Cancer Institute. Information about using the illustrations in this summary, along with many other cancer-related images, is available in Visuals Online, a collection of over 2,000 scientific images.
Disclaimer
The information in these summaries should not be used as a basis for insurance reimbursement determinations. More information on insurance coverage is available on Cancer.gov on the Managing Cancer Care page.
Contact Us
More information about contacting us or receiving help with the Cancer.gov website can be found on our Contact Us for Help page. Questions can also be submitted to Cancer.gov through the website’s Email Us.