Melanoma Treatment (PDQ®)–Health Professional Version
General Information About Melanoma
Melanoma is a malignant tumor of melanocytes, which are the cells that make the pigment melanin and are derived from the neural crest. Although most melanomas arise in the skin, they may also arise from mucosal surfaces or at other sites to which neural crest cells migrate, including the uveal tract. Uveal melanomas differ significantly from cutaneous melanoma in incidence, prognostic factors, molecular characteristics, and treatment. For more information, visit Intraocular (Uveal) Melanoma Treatment.
Incidence and Mortality
Estimated new cases and deaths from melanoma in the United States in 2025:[1]
- New cases: 104,960.
- Deaths: 8,430.
Skin cancer is the most common malignancy diagnosed in the United States. Invasive melanoma represents about 1% of skin cancers but results in the most deaths.[1,2] Since the early 2000s, the incidence of melanoma in people younger than 50 years has declined by about 1% per year in men and stabilized in women. In people aged 50 years and older, the incidence has stabilized in men and increased by about 3% per year in women.[1] Older men are at highest risk of melanoma; however, it is the most common cancer in young adults aged 25 to 29 years and the second most common cancer in those aged 15 to 29 years.[3] Ocular melanoma is the most common cancer of the eye, with approximately 2,000 cases diagnosed annually.
Risk Factors
Risk factors for melanoma include both intrinsic (genetic and phenotype) and extrinsic (environmental or exposure) factors:
- Sun exposure.
- Pigmentary characteristics.
- Multiple nevi.
- Family and personal history of melanoma.
- Immunosuppression.
- Environmental exposures.
For more information about risk factors, visit Skin Cancer Prevention and Genetics of Skin Cancer.
Anatomy
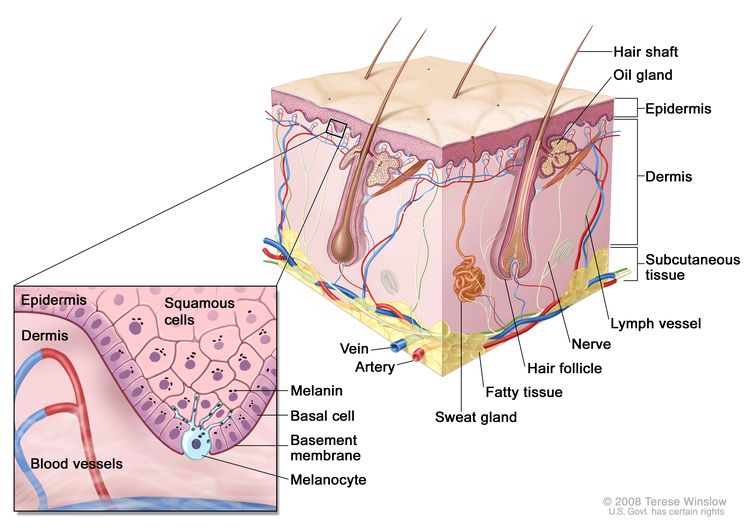
Screening
For more information, visit Skin Cancer Screening.
Clinical Features
Melanoma occurs predominantly in adults, and more than 50% of the cases arise in apparently normal areas of the skin. Melanoma can occur anywhere, including on mucosal surfaces and the uvea. However, in women it occurs more commonly on the extremities, and in men it occurs most commonly on the trunk or head and neck.[4]
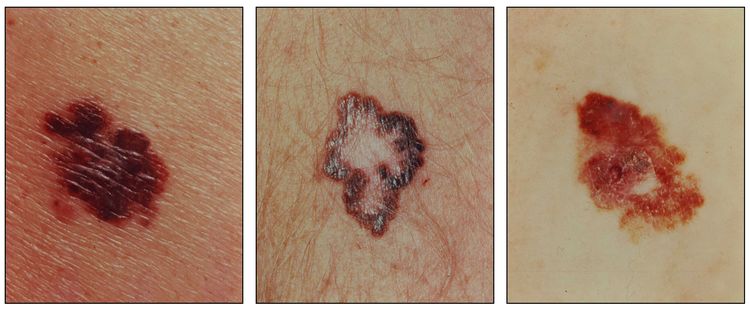
Early signs in a nevus that would suggest a malignant change include:
- Darker or variable discoloration.
- Itching.
- An increase in size or the development of satellite lesions.
- Ulceration or bleeding (later signs).
A common acronym used by medical professionals and the lay public to identify the suspicious features of pigmented lesions that may reflect malignant change is ABCDE:[5]
- Asymmetry of the lesion.
- Border irregularity.
- Color variation.
- Diameter >6 mm.
- Evolution or change in the lesion.
Diagnosis
A biopsy, preferably by local excision, should be performed for any suspicious lesions. Suspicious lesions should never be shaved off or cauterized. An experienced pathologist should examine the specimens to allow for microstaging.
Studies show that distinguishing between benign pigmented lesions and early melanomas can be difficult, and even experienced dermatopathologists can have differing opinions. To reduce the possibility of misdiagnosis for an individual patient, a second review by an independent qualified pathologist should be considered.[6,7] Agreement between pathologists in the histological diagnosis of melanomas and benign pigmented lesions has been found to be considerably variable.[6,7]
Evidence (discordance in histological evaluation):
- One study found that there was discordance in the diagnosis of melanoma versus benign lesions in 37 of 140 cases examined by a panel of experienced dermatopathologists. For the histological classification of cutaneous melanoma, the highest concordance was attained for Breslow thickness and presence of ulceration, while the agreement was poor for other histological features such as Clark level of invasion, presence of regression, and lymphocytic infiltration.[6]
- In another study, 38% of cases examined by a panel of expert pathologists had two or more discordant interpretations.[7]
Prognostic Factors
Prognosis is affected by the characteristics of primary and metastatic tumors. The most important prognostic factors have been incorporated into the 2017 eighth edition of the American Joint Committee on Cancer (AJCC) staging manual and include:[4,8-10]
- Thickness and/or level of invasion of the melanoma.
- Ulceration or bleeding at the primary site.
- Number of regional lymph nodes involved, with distinction of clinically occult and clinically apparent.
- Presence of non-nodal regional disease, including microsatellites, satellites, and in-transit cutaneous or subcutaneous metastases.
- Systemic metastasis.
- Site—nonvisceral versus lung versus all other visceral sites versus central nervous system.
- Elevated serum lactate dehydrogenase level.
Patients who are younger, female, and who have melanomas on their extremities generally have better prognoses.[4,8,9,11]
The risk of relapse decreases substantially over time, although late relapses do occur.[12-15] Long-term follow-up is important for detection of recurrence, managing long-term effects, and surveillance of new lesions.
Related Subtypes
Mucosal melanoma arises from melanocytes within the mucosal lining of the respiratory, gastrointestinal, or genitourinary tracts. This is a rare subgroup, representing only 1.4% of melanomas.[16] The etiology of mucosal melanomas remains unclear, but whole-genome sequencing reveals mutational signatures unrelated to UV radiation, which is distinct from cutaneous melanomas.[17] Diagnosis is often delayed due to the location of these lesions, a lack of symptoms, and potential amelanotic appearance.[18] There is no clear consensus on staging definitions for mucosal melanoma, and the AJCC eighth edition TNM (tumor, node, metastasis) staging is only used for head and neck mucosal melanomas.[10] The overall prognosis is poor and varies by location.
References
- American Cancer Society: Cancer Facts and Figures 2025. American Cancer Society, 2025. Available online. Last accessed January 16, 2025.
- Melanoma. Bethesda, Md: National Library of Medicine, 2012. Available online. Last accessed May 1, 2025.
- Bleyer A, O’Leary M, Barr R, et al., eds.: Cancer Epidemiology in Older Adolescents and Young Adults 15 to 29 Years of Age, Including SEER Incidence and Survival: 1975-2000. National Cancer Institute, 2006. NIH Pub. No. 06-5767. Available online. Last accessed May 1, 2025.
- Slingluff CI Jr, Flaherty K, Rosenberg SA, et al.: Cutaneous melanoma. In: DeVita VT Jr, Lawrence TS, Rosenberg SA: Cancer: Principles and Practice of Oncology. 9th ed. Lippincott Williams & Wilkins, 2011, pp 1643-91.
- Abbasi NR, Shaw HM, Rigel DS, et al.: Early diagnosis of cutaneous melanoma: revisiting the ABCD criteria. JAMA 292 (22): 2771-6, 2004. [PUBMED Abstract]
- Corona R, Mele A, Amini M, et al.: Interobserver variability on the histopathologic diagnosis of cutaneous melanoma and other pigmented skin lesions. J Clin Oncol 14 (4): 1218-23, 1996. [PUBMED Abstract]
- Farmer ER, Gonin R, Hanna MP: Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol 27 (6): 528-31, 1996. [PUBMED Abstract]
- Balch CM, Soong S, Ross MI, et al.: Long-term results of a multi-institutional randomized trial comparing prognostic factors and surgical results for intermediate thickness melanomas (1.0 to 4.0 mm). Intergroup Melanoma Surgical Trial. Ann Surg Oncol 7 (2): 87-97, 2000. [PUBMED Abstract]
- Manola J, Atkins M, Ibrahim J, et al.: Prognostic factors in metastatic melanoma: a pooled analysis of Eastern Cooperative Oncology Group trials. J Clin Oncol 18 (22): 3782-93, 2000. [PUBMED Abstract]
- Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. Springer; 2017, pp. 563–85.
- Balch CM, Gershenwald JE, Soong SJ, et al.: Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 27 (36): 6199-206, 2009. [PUBMED Abstract]
- Shen P, Guenther JM, Wanek LA, et al.: Can elective lymph node dissection decrease the frequency and mortality rate of late melanoma recurrences? Ann Surg Oncol 7 (2): 114-9, 2000. [PUBMED Abstract]
- Tsao H, Cosimi AB, Sober AJ: Ultra-late recurrence (15 years or longer) of cutaneous melanoma. Cancer 79 (12): 2361-70, 1997. [PUBMED Abstract]
- Sarac E, Wilhelmi J, Thomas I, et al.: Late recurrence of melanoma after 10 years - Is the course of the disease different from early recurrences? J Eur Acad Dermatol Venereol 34 (5): 977-983, 2020. [PUBMED Abstract]
- Faries MB, Steen S, Ye X, et al.: Late recurrence in melanoma: clinical implications of lost dormancy. J Am Coll Surg 217 (1): 27-34; discussion 34-6, 2013. [PUBMED Abstract]
- McLaughlin CC, Wu XC, Jemal A, et al.: Incidence of noncutaneous melanomas in the U.S. Cancer 103 (5): 1000-7, 2005. [PUBMED Abstract]
- Hayward NK, Wilmott JS, Waddell N, et al.: Whole-genome landscapes of major melanoma subtypes. Nature 545 (7653): 175-180, 2017. [PUBMED Abstract]
- Thomas NE, Kricker A, Waxweiler WT, et al.: Comparison of clinicopathologic features and survival of histopathologically amelanotic and pigmented melanomas: a population-based study. JAMA Dermatol 150 (12): 1306-314, 2014. [PUBMED Abstract]
Cellular and Molecular Classification of Melanoma
The descriptive terms for clinicopathological cellular subtypes of malignant melanoma are of historical interest only; they do not have independent prognostic or therapeutic significance. These cellular subtypes include:
- Superficial spreading.
- Nodular.
- Lentigo maligna.
- Acral lentiginous (palmar/plantar and subungual).
- Miscellaneous unusual types:
- Mucosal lentiginous (oral and genital).
- Desmoplastic.
- Verrucous.
Genomic Classification
Cutaneous melanoma
The Cancer Genome Atlas (TCGA) Network performed an integrative multiplatform characterization of 333 cutaneous melanomas from 331 patients.[1] Using six types of molecular analysis at the DNA, RNA, and protein levels, the researchers identified four major genomic subtypes:
- BRAF-altered.
- RAS-altered.
- NF1-altered.
- Triple wild-type.
Genomic subtypes may suggest drug targets and clinical trial design, as well as guide clinical decision-making for targeted therapies. For more information, visit Table 1.
Targeted therapies have demonstrated efficacy and received U.S. Food and Drug Administration approval for the BRAF-altered subtype of melanoma. Combination therapies with a BRAF plus a MEK inhibitor have shown improvement in outcomes over a single-agent inhibitor alone. However, virtually all patients acquire resistance to this therapy and experience disease relapse. Therefore, clinical trials remain an important option for patients with melanoma and a BRAF pathogenic variant and other genomic subtypes of melanoma. For more information, visit the individual treatment sections.
A variety of immunotherapies have been approved for the treatment of melanoma regardless of genetic subtype. The benefit of immunotherapy has not been associated with a specific pathogenic variant or molecular subtype. The TCGA analysis identified immune markers (in a subset within each molecular subtype) that were associated with improved survival and that may have implications for immunotherapy. Identification of predictive biomarkers remains an active area of research. For more information, visit the individual treatment sections.
| Genomic Subtype | % Samples With Pathogenic Variant | Increased Lymphocytic Infiltration (%) | Clinical Management Implications for Targeted Therapyb,c | ||
|---|---|---|---|---|---|
| FDA = U.S. Food and Drug Administration; WT = wild-type. | |||||
| aPrimary melanoma with matched normal samples; N = 67 (20%). Metastatic melanoma with matched normal samples; N = 266 (80%). Matched is defined as sample from the same patient. | |||||
| bThe indications for immunotherapy are not known to be determined or limited by genomic subtype. | |||||
| cRisks and benefits of single versus combination therapies are detailed in the Treatment Option Overview for Melanoma section of this summary. | |||||
| dResearch includes but is not limited to these examples. Clinical trials are posted on clinicaltrials.gov. | |||||
| eIndicated when variant is diagnosed by an FDA-approved assay. | |||||
| fTriple WT was defined as a heterogeneous subgroup lacking BRAF, NRAS, HRAS, and KRAS, and NF1 pathogenic variants. | |||||
| FDA Approved | Researchd (single agent or in combination) | ||||
| BRAF-altered | 52 | ~ 30 | BRAF inhibitorse | CDK inhibitors, PI3K/Akt/mTOR inhibitors, ERK inhibitors, IDH1 inhibitors, EZH2 inhibitors, Aurora kinase inhibitors, ARID2 chromatin remodelers | |
| – Vemurafenib | |||||
| –Dabrafenib | |||||
| –Encorafenib | |||||
| MEK inhibitors | |||||
| –Trametinib | |||||
| –Cobimetinib | |||||
| –Binimetinib | |||||
| Combination of BRAF + MEK inhibitors | |||||
| –Vemurafenib + cobimetinib | |||||
| –Dabrafenib + trametinib | |||||
| –Encorafenib + binimetinib | |||||
| RAS-altered (NRAS, HRAS, and KRAS ) | 28 | ~ 25 | MEK inhibitors, CDK inhibitors, PI3K/Akt/mTOR inhibitors, ERK inhibitors, IDH1 inhibitors, EZH2 inhibitors, Aurora kinase inhibitors, ARID2 chromatin remodelers | ||
| NF1-altered | 14 | ~ 25 | PI3K/Akt/mTOR inhibitors, ERK inhibitors, IDH1 inhibitors, EZH2 inhibitors, ARID2 chromatin remodelers | ||
| Triple WTf | 14.5 | ~ 40 | KIT-altered/amplified CDK inhibitors (i.e., imatinib and dasatinib), MDM2/p53 interaction inhibitors, PI3K/Akt/mTOR inhibitors, IDH1 inhibitors, EZH2 inhibitors | ||
Uveal melanoma
Uveal melanomas differ significantly from cutaneous melanomas. ln one series, 83% of 186 uveal melanomas were found to have a constitutively active somatic pathogenic variant in GNAQ or GNA11.[2,3] For more information, visit Intraocular (Uveal) Melanoma Treatment.
References
- Cancer Genome Atlas Network: Genomic Classification of Cutaneous Melanoma. Cell 161 (7): 1681-96, 2015. [PUBMED Abstract]
- Van Raamsdonk CD, Bezrookove V, Green G, et al.: Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457 (7229): 599-602, 2009. [PUBMED Abstract]
- Van Raamsdonk CD, Griewank KG, Crosby MB, et al.: Mutations in GNA11 in uveal melanoma. N Engl J Med 363 (23): 2191-9, 2010. [PUBMED Abstract]
Stage Information for Melanoma
Clinical staging is based on the thickness and ulceration status of the primary tumor, and whether the tumor has spread to regional lymph nodes or distant sites. For melanoma that is clinically confined to the primary site, the chance of lymph node or systemic metastases increases as the thickness and depth of local invasion increases, which worsens the prognosis. Melanoma can spread by local extension (through lymphatics) and/or by hematogenous routes to distant sites. Any organ may be involved by metastases, but the lungs and liver are common sites.
The microstage of malignant melanoma is determined on histological examination by the vertical thickness of the lesion in millimeters (Breslow classification) and/or the anatomical level of local invasion (Clark classification). The Breslow thickness is more reproducible and more accurately predicts subsequent behavior of malignant melanoma in lesions thicker than 1.5 mm and should always be reported.
Accurate microstaging of the primary tumor requires careful histological evaluation of the entire specimen by an experienced pathologist.
Clark Classification (Level of Invasion)
| Level of Invasion | Description |
|---|---|
| Level I | Lesions involving only the epidermis (in situ melanoma); not an invasive lesion. |
| Level II | Invasion of the papillary dermis; does not reach the papillary-reticular dermal interface. |
| Level III | Invasion fills and expands the papillary dermis but does not penetrate the reticular dermis. |
| Level IV | Invasion into the reticular dermis but not into the subcutaneous tissue. |
| Level V | Invasion through the reticular dermis into the subcutaneous tissue. |
AJCC Stage Groupings and TNM Definitions
The American Joint Committee on Cancer (AJCC) has designated staging by TNM (tumor, node, metastasis) classification to define melanoma.[1]
Cancers staged using this staging system include cutaneous melanoma. Cancers not staged using this system include melanoma of the conjunctiva; melanoma of the uvea; mucosal melanoma arising in the head and neck; mucosal melanoma of the urethra, vagina, rectum, and anus; Merkel cell carcinoma; and squamous cell carcinoma.[1]
AJCC Prognostic Stage Groups-Clinical (cTNM)
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) |
|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; c = clinical; LDH = lactate dehydrogenase; No. = number. | ||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | ||||
| bThickness and ulceration status not applicable. | ||||
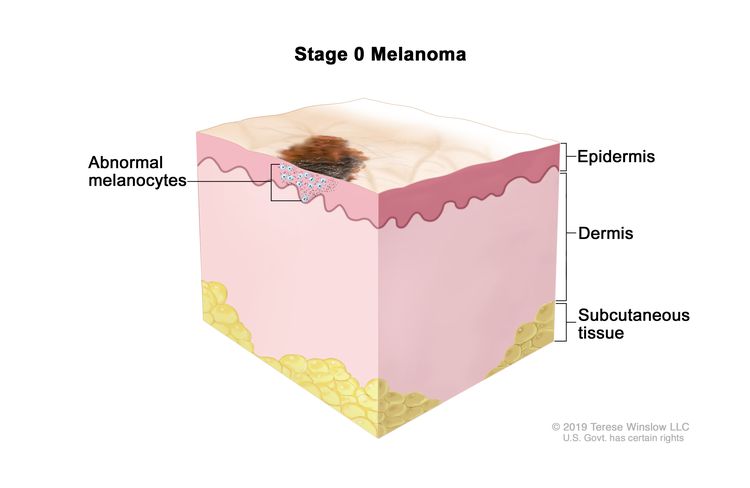
| 0 | Tis, N0, M0 | Tis = Melanoma in situ.b | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) |
|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; c = clinical; LDH = lactate dehydrogenase; No. = number. | ||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | ||||
| IA | T1a, N0, M0 | T1a = <0.8 mm/without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
| IB | T1b, N0, M0 | T1b = <0.8 mm with ulceration; 0.8–1.0 mm with or without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
| T2a, N0, M0 | T2a = >1.0–2.0 mm/without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. | |
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) |
|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; c = clinical; LDH = lactate dehydrogenase; No. = number. | ||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | ||||
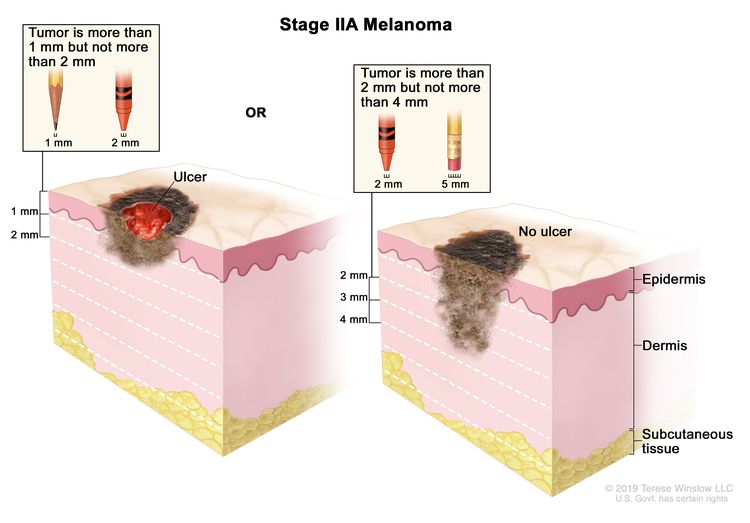
| IIA | T2b, N0, M0 | T2b = >1.0–2.0 mm/with ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
| T3a, N0, M0 | T3a = >2.0–4.0 mm/without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. | |
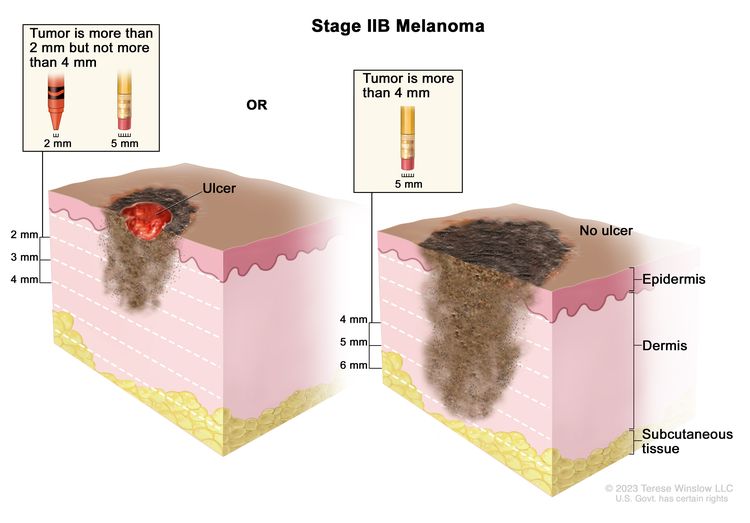
| IIB | T3b, N0, M0 | T3b = >2.0–4.0 mm/with ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
| T4a, N0, M0 | T4a = >4.0 mm/without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. | |
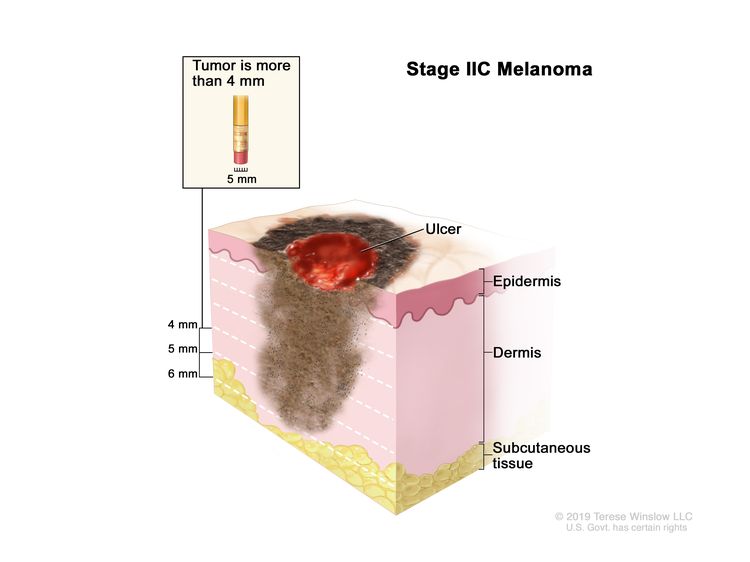
| IIC | T4b, N0, M0 | T4b = >4.0 mm/with ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH) |
|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; c = clinical; LDH = lactate dehydrogenase; No. = number. | ||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | ||||
| bFor example, diagnosis by curettage. | ||||
| cFor example, unknown primary or completely regressed melanoma. | ||||
| dThickness and ulceration status not applicable. | ||||
| eDetected by sentinel lymph node biopsy. | ||||
| III | Any T, Tis, ≥N1, M0 | TX = Primary tumor cannot be assessed.b,d | N1a = One clinically occult nodee /in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. |
| T0 = No evidence of primary tumor.c,d | ||||
| Tis = Melanoma in situ.d | N1b = One clinically detected node/in-transit, satellite, and/or microsatellite metastases not present. | |||
| T1a = <0.8 mm/without ulceration. | N1c = No regional lymph node disease/in-transit, satellite, and/or microsatellite metastases present. | |||
| T1b = <0.8 mm with ulceration; 0.8–1.0 mm with or without ulceration. | N2a = Two or three clinically occult nodese/in-transit, satellite, and/or microsatellite metastases not present. | |||
| T2a = >1.0–2.0 mm/without ulceration. | N2b = Two or three nodes at least one of which was clinically detected/in-transit, satellite, and or microsatellite metastases not present. | |||
| T2b = >1.0–2.0 mm/with ulceration. | N2c = One clinically occult or clinically detected node/in-transit, satellite, and/or microsatellite metastases present. | |||
| T3a = >2.0–4.0 mm/without ulceration. | N3a = Four or more clinically occult nodese/in-transit, satellite, and/or microsatellite metastases not present. | |||
| T3b = >2.0–4.0 mm/with ulceration. | N3b = Four or more nodes, at least one of which was clinically detected, or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases not present. | |||
| T4a = >4.0 mm/without ulceration. | N3c = Two or more clinically occult or clinically detected nodes and/or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases present. | |||
| T4b = >4.0 mm/with ulceration. | ||||
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) |
|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; c = clinical; CNS = central nervous system; LDH = lactate dehydrogenase; No. = number. | ||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | ||||
| bFor example, sentinel lymph node biopsy not performed, regional nodes previously removed for another reason. (Exception: pathological N category is not required for T1 melanomas, use cN). | ||||
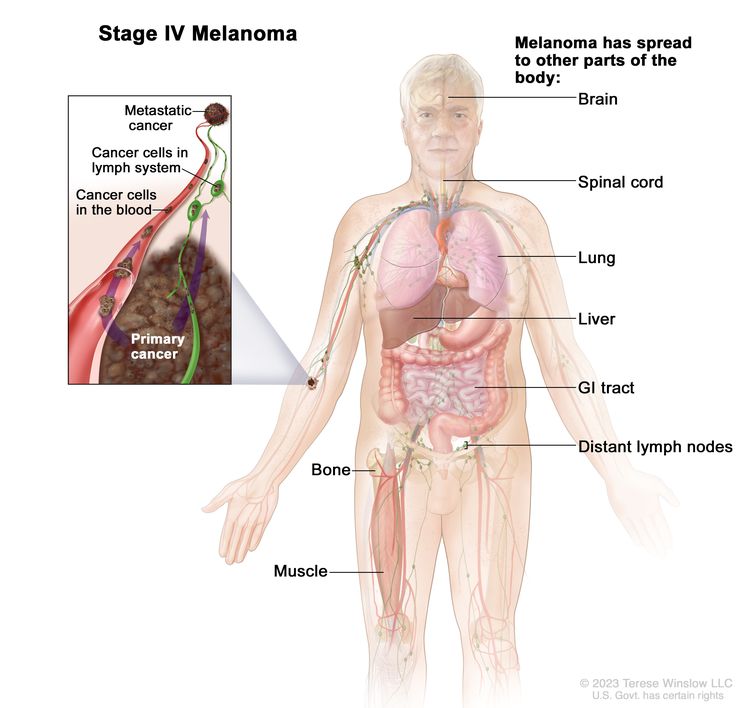
| IV | Any T, Any N, M1 | Any T = Visit Table 6 for description. | NX = Regional nodes not assessed;b N0 = No regional metastases; ≥N1 = Visit Table 6 for description. | M1 = Evidence of distant metastasis. |
| –M1a = Distant metastasis to skin, soft tissue including muscle, and/or nonregional lymph nodes [M1a(0) = LDH not elevated; M1a(1) = LDH elevated]. | ||||
| –M1b = Distant metastasis to lung with or without M1a sites of disease [M1b(0) = LDH not elevated; M1b(1) = LDH elevated]. | ||||
| –M1c = Distant metastasis to non-CNS visceral sites with or without M1a or M1b sites of disease [M1c(0) = LDH not elevated; OR M1c(1) = LDH elevated]. | ||||
| –M1d = Distant metastasis to CNS with or without M1a, M1b, or M1c sites of disease [M1d(0) = LDH not elevated; M1d(1) = LDH elevated]. | ||||
AJCC Prognostic Stage Groups-Pathological (pTNM)
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) | Illustration |
|---|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; cN = clinical N; LDH = lactate dehydrogenase; No. = number; p = pathological. | |||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | |||||
| bPathological stage 0 (melanoma in situ) and T1 do not require pathological evaluation of lymph nodes to complete pathological staging; use cN information to assign their pathological stage. | |||||
| cThickness and ulceration status not applicable. | |||||
| 0 | Tis, N0, M0 | Tis = Melanoma in situ.b,c | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
 |
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) | Illustration |
|---|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; cN = clinical N; LDH = lactate dehydrogenase; No. = number; p = pathological. | |||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | |||||
| bPathological stage 0 (melanoma in situ) and T1 do not require pathological evaluation of lymph nodes to complete pathological staging; use cN information to assign their pathological stage. | |||||
| IA | T1a, N0, M0 | T1a = <0.8 mm/without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
 |
| T1b, N0, M0 | T1b = <0.8 mm with ulceration; 0.8–1.0 mm with or without ulceration. | ||||
| IB | T2a, N0, M0 | T2a = >1.0–2.0 mm/without ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. | |
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) | Illustration |
|---|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; LDH = lactate dehydrogenase; No. = number; p = pathological. | |||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | |||||
| IIA | T2b, N0, M0 | T2b = >1.0–2.0 mm/with ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
 |
| T3a, N0, M0 | T3a = >2.0–4.0 mm/without ulceration. | ||||
| IIB | T3b, N0, M0 | T3b = >2.0–4.0 mm/with ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
 |
| T4a, N0, M0 | T4a = >4.0 mm/without ulceration. | ||||
| IIC | T4b, N0, M0 | T4b = >4.0 mm/with ulceration. | N0 = No regional metastases detected. | M0 = No evidence of distant metastasis. |
 |
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) |
|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; LDH = lactate dehydrogenase; No. = number; p = pathological. | ||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | ||||
| bDetected by sentinel lymph node biopsy. | ||||
| cFor example, unknown primary or completely regressed melanoma. | ||||
| dThickness and ulceration status not applicable. | ||||
| IIIA | T1a/b–T2a, N1a or N2a, M0 | T1a = <0.8 mm/without ulceration/T1b = <0.8 mm with ulceration; 0.8–1.0 mm with or without ulceration. | N1a = One clinically occult nodeb/in-transit, satellite, and/or microsatellite metastases not present; OR N2a = Two or three clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. |
| T2a = >1.0–2.0 mm/without ulceration. | ||||
| IIIB | T0, N1b, N1c, M0 | T0 = No evidence of primary tumor.c,d | N1b = One clinically detected node/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. |
| N1c = No regional lymph node disease/in-transit, satellite, and/or microsatellite metastases present. | ||||
| T1a/b–T2a, N1b/c or N2b, M0 | T1a = <0.8 mm/without ulceration/T1b <0.8 mm with ulceration; 0.8–1.0 mm with or without ulceration. | N1b = One clinically detected node/in-transit, satellite, and/or microsatellite metastases not present;/N1c = No regional lymph node disease/in-transit, satellite, and/or microsatellite metastases present; OR | M0 = No evidence of distant metastasis. | |
| T2a = >1.0–2.0 mm/without ulceration. | ||||
| N2b = Two or three nodes at least one of which was clinically detected/in-transit, satellite, and or microsatellite metastases not present. | ||||
| T2b/T3a, N1a–N2b, M0 | T2b = >1.0–2.0 mm/with ulceration/T3a = >2.0–4.0 mm/without ulceration. | N1a = One clinically occult nodeb/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. | |
| N1b = One clinically detected node/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N1c = No regional lymph node disease/in-transit, satellite, and/or microsatellite metastases present. | ||||
| N2a = Two or three clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N2b = Two or three nodes, at least one of which was clinically detected/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| IIIC | T0, N2b, N2c, N3b, or N3c, M0 | T0 = No evidence of primary tumor.c,d | N2b = Two or three nodes, at least one of which was clinically detected/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. |
| N2c = One clinically occult or clinically detected node/in-transit, satellite, and/or microsatellite metastases present. | ||||
| N3b = Four or more nodes, at least one of which was clinically detected, or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases not present; OR | ||||
| N3c = Two or more clinically occult or clinically detected nodes and/or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases present. | ||||
| T1a–T3a, N2c or N3a/b/c, M0 | T1a = <0.8 mm/without ulceration/T1b = <0.8 mm with ulceration; 0.8–1.0 mm with or without ulceration. | N2c = One clinically occult or clinically detected node/in-transit, satellite, and/or microsatellite metastases present; OR | M0 = No evidence of distant metastasis. | |
| T2a = >1.0–2.0 mm/without ulceration. | ||||
| T2b = >1.0–2.0 mm/with ulceration. | ||||
| N3a = Four or more clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N3b = Four or more nodes, at least one of which was clinically detected, or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| T3a = >2.0–4.0 mm/without ulceration. | N3c = Two or more clinically occult or clinically detected nodes and/or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases present. | |||
| T3b/T4a, Any N ≥N1, M0 | T3b = >2.0–4.0 mm/with ulceration/T4a = >4.0 mm/without ulceration. | N1a = One clinically occult nodeb/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. | |
| N1b = One clinically detected node/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N1c = No regional lymph node disease/in-transit, satellite, and/or microsatellite metastases present. | ||||
| N2a = Two or three clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N2b = Two or three nodes, at least one of which was clinically detected/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N2c = One clinically occult or clinically detected node/in-transit, satellite, and/or microsatellite metastases present. | ||||
| N3a = Four or more clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N3b = Four or more nodes, at least one of which was clinically detected, or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N3c = Two or more clinically occult or clinically detected nodes and/or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases present. | ||||
| T4b, N1a–N2c, M0 | T4b = >4.0 mm/with ulceration. | N1a = One clinically occult nodeb/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. | |
| N1b = One clinically detected node/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N1c = No regional lymph node disease/in-transit, satellite, and/or microsatellite metastases present. | ||||
| N2a = Two or three clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N2b = Two or three nodes, at least one of which was clinically detected/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N2c = One clinically occult or clinically detected node/in-transit, satellite, and/or microsatellite metastases present. | ||||
| IIID | T4b, N3a/b/c, M0 | T4b = >4.0 mm/with ulceration. | N3a = Four or more clinically occult nodesb/in-transit, satellite, and/or microsatellite metastases not present. | M0 = No evidence of distant metastasis. |
| N3b = Four or more nodes, at least one of which was clinically detected, or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases not present. | ||||
| N3c = Two or more clinically occult or clinically detected nodes and/or presence of any number of matted nodes/in-transit, satellite, and/or microsatellite metastases present. | ||||
| Stage | TNM | T Category (Thickness/Ulceration Status) | N Category (No. of Tumor-Involved Regional Lymph Nodes/Presence of In-Transit, Satellite, and/or Microsatellite Metastases) | M Category (Anatomic Site/LDH Level) | Illustration |
|---|---|---|---|---|---|
| T = primary tumor; N = regional lymph node; M = distant metastasis; cN = clinical N; CNS = central nervous system; LDH = lactate dehydrogenase; No. = number; p = pathological. | |||||
| aAdapted from AJCC: Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer, 2017, pp. 563–85. | |||||
| bFor example, sentinel lymph node biopsy not performed, regional nodes previously removed for another reason. (Exception: pathological N category is not required for T1 melanomas, use cN). | |||||
| cPathological stage 0 (melanoma in situ) and T1 do not require pathological evaluation of lymph nodes to complete pathological staging; use cN information to assign their pathological stage. | |||||
| dThickness and ulceration status not applicable. | |||||
| IV | Any T, Tis, Any N, M1 | Any T = Visit Table 6 for description. | NX = Regional nodes not assessed;d N0 = No regional metastases; ≥N1 = Visit Table 6 for description. | M1 = Evidence of distant metastasis. |
 |
| Tis = Melanoma in situ.b,c | –M1a = Distant metastasis to skin, soft tissue including muscle, and/or nonregional lymph nodes [M1a(0) = LDH not elevated; M1a(1) = LDH elevated]. | ||||
| –M1b = Distant metastasis to lung with or without M1a sites of disease [M1b(0) = LDH not elevated; M1b(1) = LDH elevated]. | |||||
| –M1c - Distant metastasis to non-CNS visceral sites with or without M1a or M1b sites of disease [M1c(0) = LDH not elevated; M1c(1) = LDH elevated]. | |||||
| –M1d = Distant metastasis to CNS with or without M1a, M1b, or M1c sites of disease [M1d(0) = LDH not elevated; M1d(1) = LDH elevated]. | |||||
References
- Melanoma of the Skin. In: Amin MB, Edge SB, Greene FL, et al., eds.: AJCC Cancer Staging Manual. 8th ed. Springer; 2017, pp. 563–85.
Treatment Option Overview for Melanoma
| Stage (TNM Staging Criteria) | Treatment Optionsa |
|---|---|
| aClinical trials are an important option for patients with all stages of melanoma because advances in understanding the aberrant molecular and biological pathways have led to rapid drug development. Standard treatment options are available in many clinical trials. Information about ongoing clinical trials is available from the NCI website. | |
| Stage 0 melanoma | Excision |
| Stage IA melanoma | Excision +/− sentinel lymph node biopsy |
| Stage IB melanoma | Excision with lymph node management |
| Stage II melanoma | Excision with lymph node management |
| Adjuvant therapy | |
| Resectable Stage III melanoma | Excision +/− lymph node management |
| Neoadjuvant therapy | |
| Adjuvant therapy | |
| Combination immunotherapies, including vaccines (under clinical evaluation) | |
| Adjuvant therapies that target a known pathogenic variant, e.g., KIT (under clinical evaluation) | |
| Intralesional therapies (under clinical evaluation) | |
| Unresectable Stage III, Stage IV, and Recurrent melanoma | Immunotherapy |
| Signal transduction inhibitors | |
| Intralesional therapy | |
| Adjunctive local/regional therapy including surgical resection | |
| Palliative therapy | |
| Targeted therapy with single agents or combination therapy (under clinical evaluation) | |
| Combinations of immunotherapy and targeted therapy (under clinical evaluation) | |
| Intralesional injections (e.g., oncolytic viruses) (under clinical evaluation) | |
| Complete surgical resection of all known disease versus best medical therapy (under clinical evaluation) | |
| Isolated limb perfusion for unresectable extremity melanoma (under clinical evaluation) | |
| Systemic therapy for unresectable disease (under clinical evaluation) | |
Excision
Surgical excision remains the primary modality for treating localized melanoma. Cutaneous melanomas that have not spread beyond the initial site are highly curable. Localized melanoma is excised with margins proportional to the microstage of the primary lesion.
Standardizing treatment for mucosal melanoma is difficult due to the paucity of prospective data in this rare subgroup. Surgery remains the cornerstone of therapy. Local excision is performed when feasible, as radical resection has not conferred a survival advantage in retrospective studies.[1-3] Optimal excision margins have not been prospectively studied, but the ultimate goal is to obtain negative histological margins.[4]
Lymph node management
Sentinel lymph node biopsy (SLNB)
Lymphatic mapping and SLNB should be considered to assess the presence of occult metastasis in the regional lymph nodes of patients with primary tumors measuring at least 0.8 mm thick with clinically negative nodes. These procedures may identify individuals who can avoid regional lymph node dissection and individuals who may benefit from adjuvant therapy.[5-10]
Multiple studies have demonstrated the diagnostic accuracy of SLNB, with false-negative rates of 0% to 2%.[5,10-15] To ensure accurate identification of the sentinel lymph node, lymphatic mapping and removal of the sentinel lymph node are ideally performed during the same operation as the wide excision of the primary melanoma.
If micrometastatic melanoma is detected, active surveillance with ultrasound of the draining nodal basin is an acceptable treatment recommendation that has widely replaced complete lymph node dissection (CLND). A complete regional lymphadenectomy can be considered in select populations.
For clinically node-negative patients, there is insufficient evidence to define the role of SLNB in sinonasal, anorectal, or vaginal melanoma. However, SLNB has shown feasibility and accuracy in vulvar melanoma.[16,17] Therapeutic dissection is indicated for clinically positive regional nodes in the absence of distant disease.
Complete lymph node dissection (CLND)
Patients can consider CLND for regional control if the sentinel node(s) is microscopically or macroscopically positive.
Adjuvant Therapy
Adjuvant therapy options for patients at high risk of recurrence after complete resection include checkpoint inhibitors and combination signal transduction inhibitor therapy. Ipilimumab was the first checkpoint inhibitor to be approved by the U.S. Food and Drug Administration (FDA) as adjuvant therapy. It has demonstrated improved overall survival (OS) when given at 10 mg/kg (ipi10), compared with placebo (EORTC 18071 [NCT00636168]).[18] However, ipi10 has significant toxicity at this dose. The North American Intergroup Trial E1609 (NCT01274338), designed with three treatment groups, compared ipi10 with ipilimumab at a lower dose of 3 mg/kg (ipi3) (approved for metastatic melanoma) and with high-dose interferon (HDI). Patients who received ipi3 had a significant improvement in OS compared with ipi10.[19] These data do not support HDI as an adjuvant treatment option for melanoma.
Large randomized trials with nivolumab and pembrolizumab and with combination signal transduction inhibitors (dabrafenib plus trametinib) have shown a clinically significant impact on relapse-free survival (RFS). CheckMate 238 (NCT02388906) compared nivolumab with ipi10 and found that nivolumab produced superior RFS and had a more tolerable safety profile.[20] Pembrolizumab produced superior RFS compared with placebo, with data on OS still maturing in the MK-3475-054/KEYNOTE-054 trial (NCT02362594).[21] Dabrafenib plus trametinib produced superior RFS compared with placebo, with data on OS still maturing in the COMBI-AD trial (NCT01682083).[22] Single-agent BRAF-inhibitor therapy with vemurafenib did not show improved RFS compared with placebo in the BRIM8 trial (NCT01667419).[23]
The benefit of immunotherapy with ipilimumab, nivolumab, and pembrolizumab has been seen regardless of programmed death-ligand 1 (PD-L1) expression or the presence of BRAF pathogenic variants. Combination signal transduction inhibitor therapy is an additional option for patients with BRAF variants.
Participation in clinical trials designed to identify treatments that will further extend RFS and OS with less toxicity and shorter treatment schedules is an important option for all patients.
Neoadjuvant Therapy
Neoadjuvant pembrolizumab can be considered for patients with high-risk node-positive disease or resectable metastatic disease based on the results of a phase II trial (SWOG S1801 [NCT03698019]). In the trial, patients who received 1 year of neoadjuvant and adjuvant pembrolizumab had improved event-free survival (EFS) compared with those who underwent primary resection and received adjuvant pembrolizumab.[24] A total of 313 patients with resectable macroscopic stage IIIB to stage IV melanoma were randomly assigned to receive either (1) three cycles of neoadjuvant pembrolizumab (200 mg intravenously [IV] every 3 weeks) followed by surgery and fifteen cycles of adjuvant pembrolizumab or (2) primary surgery followed by eighteen cycles of adjuvant pembrolizumab (200 mg IV every 3 weeks).
At a median follow-up of 15 months, the EFS rate was 72% in patients who received neoadjuvant immunotherapy and 49% in patients who received adjuvant therapy. Neoadjuvant pembrolizumab did not significantly increase toxicity in the perioperative period compared with adjuvant therapy. OS data are not available yet. The FDA has not approved this regimen, pending the completion of a randomized phase III trial.
Systematic Treatment for Unresectable Stage III, Stage IV, and Recurrent Disease
Treatment options for patients with metastatic melanoma have rapidly expanded over the last decade. Two approaches—checkpoint inhibition and targeting the mitogen-activated protein kinase pathway—have improved OS in randomized trials. Given the rapid development of new agents and combinations, patients may consider a clinical trial for initial treatment and at the time of any subsequent progression.
Immunotherapy
Checkpoint inhibitors
Pembrolizumab, nivolumab, ipilimumab, and relatlimab (in a fixed-dose formulation with nivolumab) are checkpoint inhibitors approved by the FDA. Each has demonstrated the ability to impact OS against different comparators in unresectable or advanced disease. Multiple phase III trials are in progress to determine the optimal sequencing of immunotherapies, immunotherapy with targeted therapy, and whether combinations of immunotherapies or immunotherapy plus targeted therapy are superior for increasing OS.
Interleukin-2 (IL-2)
The FDA approved IL-2 in 1998 because of durable complete response rates in a minority of patients (6%–7%) with previously treated metastatic melanoma in eight phase I and II studies. Phase III trials have not been conducted to compare high-dose IL-2 with other treatments or to determine the impact on OS.
Dual checkpoint inhibition
The combination of an anti–programmed death-1 (PD-1) antibody and an anti–cytotoxic T-lymphocyte antigen-4 (CTLA-4) antibody (nivolumab and ipilimumab) has prolonged progression-free survival (PFS) and OS compared with ipilimumab monotherapy. However, this combination is associated with significant toxicity.
Studies have demonstrated a PFS benefit for patients who receive the combination of nivolumab and the anti–lymphocyte-activation gene-3 (LAG-3) antibody relatlimab, compared with nivolumab monotherapy.[25] OS and response rate data are immature.
Signal transduction inhibitors
Studies indicate that both BRAF and MEK inhibitors can significantly impact the natural history of melanoma, although they do not appear to be curative as single agents. Three combination regimens of BRAF and MEK inhibitors have improved PFS and OS compared with BRAF inhibitor monotherapy.
BRAF inhibitors
Vemurafenib
Vemurafenib, approved by the FDA in 2011, has improved PFS and OS in patients with unresectable or advanced disease. Vemurafenib is an orally available, small-molecule, selective BRAF V600E kinase inhibitor, and its indication is limited to patients with a demonstrated BRAF V600E pathogenic variant by an FDA-approved test.[15]
Dabrafenib
Dabrafenib is an orally available, small-molecule, selective BRAF inhibitor that was approved by the FDA in 2013. An international multicenter trial (BREAK-3 [NCT01227889]) showed that dabrafenib improved PFS when compared with dacarbazine.[26]
Encorafenib
Encorafenib is an orally available, small-molecule, selective BRAF inhibitor. The FDA approved encorafenib in 2018 in combination with the MEK inhibitor binimetinib. A phase III randomized study demonstrated that encorafenib improved PFS and OS when compared with vemurafenib monotherapy.[27]
MEK inhibitors
Trametinib
Trametinib is an orally available, small-molecule, selective inhibitor of MEK1 and MEK2. The FDA approved trametinib in 2013 for patients with unresectable or metastatic melanoma with BRAF V600E or V600K pathogenic variants. Trametinib demonstrated improved PFS when compared with dacarbazine.[28]
Cobimetinib
Cobimetinib is an orally available, small-molecule, selective MEK inhibitor. The FDA approved cobimetinib in 2015 for use in combination with the BRAF inhibitor vemurafenib. For more information, visit the Combination signal transduction inhibitor therapy section.
Binimetinib
Binimetinib is an orally available, small-molecule, selective MEK1 and MEK2 inhibitor. The FDA approved binimetinib in 2018 for use in combination with the BRAF inhibitor encorafenib.
c-KIT inhibitors
Early data suggest that mucosal or acral melanomas with activating pathogenic variants or amplifications in KIT may be sensitive to a variety of c-KIT inhibitors.[29-31] Phase II and phase III trials are available for patients with unresectable stage III or stage IV melanoma and a KIT pathogenic variant.
Combination signal transduction inhibitor therapy
The FDA approved the combination regimens dabrafenib plus trametinib, vemurafenib plus cobimetinib, and encorafenib plus binimetinib in patients with unresectable or metastatic melanomas that carry BRAF V600E or V600K pathogenic variants as confirmed by an FDA-approved test. The approvals were based on improved PFS and OS when compared with a single-agent BRAF inhibitor (either dabrafenib or vemurafenib).
Combination signal transduction inhibitor therapy plus anti–PD-L1 therapy
The triplet regimen of cobimetinib (MEK inhibitor), vemurafenib (BRAF kinase inhibitor), and atezolizumab (PD-L1 inhibitor) is an FDA-approved regimen. A phase III study showed improved PFS over the combination of cobimetinib and vemurafenib.[32] However, there was not a significant difference in OS between the arms of the study and there was a higher rate of toxicities in the combination arm. As such, this regimen is not commonly used.
Chemotherapy
Dacarbazine
Dacarbazine was approved in 1970 based on overall response rates. Phase III trials indicated an overall response rate of 10% to 20%, with rare complete responses observed. An impact on OS has not been demonstrated in randomized trials.[33-36] When used as a control arm for registration trials of ipilimumab and vemurafenib in previously untreated patients with metastatic melanoma, dacarbazine was shown to be inferior for OS.
Temozolomide
Temozolomide, an oral alkylating agent, appeared to be similar to IV dacarbazine in a randomized phase III trial with a primary end point of OS. However, because the trial was designed to demonstrate the superiority of temozolomide, which was not achieved, the trial was left with a sample size that was inadequate to provide statistical proof of noninferiority.[34]
Palliative local therapy
Regional lymphadenectomy may be used as palliative care for melanoma that is metastatic to distant, lymph node–bearing areas. Resection may be used as palliative care for isolated metastases to the lung, gastrointestinal tract, bone, or sometimes the brain, with occasional long-term survival.[20,37,38]
References
- Moreno MA, Roberts DB, Kupferman ME, et al.: Mucosal melanoma of the nose and paranasal sinuses, a contemporary experience from the M. D. Anderson Cancer Center. Cancer 116 (9): 2215-23, 2010. [PUBMED Abstract]
- Iddings DM, Fleisig AJ, Chen SL, et al.: Practice patterns and outcomes for anorectal melanoma in the USA, reviewing three decades of treatment: is more extensive surgical resection beneficial in all patients? Ann Surg Oncol 17 (1): 40-4, 2010. [PUBMED Abstract]
- Sugiyama VE, Chan JK, Shin JY, et al.: Vulvar melanoma: a multivariable analysis of 644 patients. Obstet Gynecol 110 (2 Pt 1): 296-301, 2007. [PUBMED Abstract]
- Nilsson PJ, Ragnarsson-Olding BK: Importance of clear resection margins in anorectal malignant melanoma. Br J Surg 97 (1): 98-103, 2010. [PUBMED Abstract]
- Shen P, Wanek LA, Morton DL: Is adjuvant radiotherapy necessary after positive lymph node dissection in head and neck melanomas? Ann Surg Oncol 7 (8): 554-9; discussion 560-1, 2000. [PUBMED Abstract]
- Hochwald SN, Coit DG: Role of elective lymph node dissection in melanoma. Semin Surg Oncol 14 (4): 276-82, 1998. [PUBMED Abstract]
- Wagner JD, Gordon MS, Chuang TY, et al.: Current therapy of cutaneous melanoma. Plast Reconstr Surg 105 (5): 1774-99; quiz 1800-1, 2000. [PUBMED Abstract]
- Cascinelli N, Morabito A, Santinami M, et al.: Immediate or delayed dissection of regional nodes in patients with melanoma of the trunk: a randomised trial. WHO Melanoma Programme. Lancet 351 (9105): 793-6, 1998. [PUBMED Abstract]
- Koops HS, Vaglini M, Suciu S, et al.: Prophylactic isolated limb perfusion for localized, high-risk limb melanoma: results of a multicenter randomized phase III trial. European Organization for Research and Treatment of Cancer Malignant Melanoma Cooperative Group Protocol 18832, the World Health Organization Melanoma Program Trial 15, and the North American Perfusion Group Southwest Oncology Group-8593. J Clin Oncol 16 (9): 2906-12, 1998. [PUBMED Abstract]
- Wong SL, Balch CM, Hurley P, et al.: Sentinel lymph node biopsy for melanoma: American Society of Clinical Oncology and Society of Surgical Oncology joint clinical practice guideline. J Clin Oncol 30 (23): 2912-8, 2012. [PUBMED Abstract]
- Kirkwood JM, Strawderman MH, Ernstoff MS, et al.: Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J Clin Oncol 14 (1): 7-17, 1996. [PUBMED Abstract]
- Kirkwood JM, Ibrahim JG, Sondak VK, et al.: High- and low-dose interferon alfa-2b in high-risk melanoma: first analysis of intergroup trial E1690/S9111/C9190. J Clin Oncol 18 (12): 2444-58, 2000. [PUBMED Abstract]
- Eggermont AM, Suciu S, Santinami M, et al.: Adjuvant therapy with pegylated interferon alfa-2b versus observation alone in resected stage III melanoma: final results of EORTC 18991, a randomised phase III trial. Lancet 372 (9633): 117-26, 2008. [PUBMED Abstract]
- Hancock BW, Wheatley K, Harris S, et al.: Adjuvant interferon in high-risk melanoma: the AIM HIGH Study--United Kingdom Coordinating Committee on Cancer Research randomized study of adjuvant low-dose extended-duration interferon Alfa-2a in high-risk resected malignant melanoma. J Clin Oncol 22 (1): 53-61, 2004. [PUBMED Abstract]
- Chapman PB, Hauschild A, Robert C, et al.: Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364 (26): 2507-16, 2011. [PUBMED Abstract]
- Leitao MM, Cheng X, Hamilton AL, et al.: Gynecologic Cancer InterGroup (GCIG) consensus review for vulvovaginal melanomas. Int J Gynecol Cancer 24 (9 Suppl 3): S117-22, 2014. [PUBMED Abstract]
- Trifirò G, Travaini LL, Sanvito F, et al.: Sentinel node detection by lymphoscintigraphy and sentinel lymph node biopsy in vulvar melanoma. Eur J Nucl Med Mol Imaging 37 (4): 736-41, 2010. [PUBMED Abstract]
- Eggermont AM, Chiarion-Sileni V, Grob JJ, et al.: Prolonged Survival in Stage III Melanoma with Ipilimumab Adjuvant Therapy. N Engl J Med 375 (19): 1845-1855, 2016. [PUBMED Abstract]
- Tarhini AA, Lee SJ, Hodi FS, et al.: Phase III Study of Adjuvant Ipilimumab (3 or 10 mg/kg) Versus High-Dose Interferon Alfa-2b for Resected High-Risk Melanoma: North American Intergroup E1609. J Clin Oncol 38 (6): 567-575, 2020. [PUBMED Abstract]
- Leo F, Cagini L, Rocmans P, et al.: Lung metastases from melanoma: when is surgical treatment warranted? Br J Cancer 83 (5): 569-72, 2000. [PUBMED Abstract]
- Eggermont AMM, Blank CU, Mandala M, et al.: Adjuvant Pembrolizumab versus Placebo in Resected Stage III Melanoma. N Engl J Med 378 (19): 1789-1801, 2018. [PUBMED Abstract]
- Long GV, Hauschild A, Santinami M, et al.: Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med 377 (19): 1813-1823, 2017. [PUBMED Abstract]
- Maio M, Lewis K, Demidov L, et al.: Adjuvant vemurafenib in resected, BRAFV600 mutation-positive melanoma (BRIM8): a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol 19 (4): 510-520, 2018. [PUBMED Abstract]
- Patel SP, Othus M, Chen Y, et al.: Neoadjuvant-Adjuvant or Adjuvant-Only Pembrolizumab in Advanced Melanoma. N Engl J Med 388 (9): 813-823, 2023. [PUBMED Abstract]
- Tawbi HA, Schadendorf D, Lipson EJ, et al.: Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N Engl J Med 386 (1): 24-34, 2022. [PUBMED Abstract]
- Hauschild A, Grob JJ, Demidov LV, et al.: Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380 (9839): 358-65, 2012. [PUBMED Abstract]
- Dummer R, Ascierto PA, Gogas HJ, et al.: Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 19 (10): 1315-1327, 2018. [PUBMED Abstract]
- Flaherty KT, Robert C, Hersey P, et al.: Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367 (2): 107-14, 2012. [PUBMED Abstract]
- Hodi FS, Friedlander P, Corless CL, et al.: Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol 26 (12): 2046-51, 2008. [PUBMED Abstract]
- Guo J, Si L, Kong Y, et al.: Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol 29 (21): 2904-9, 2011. [PUBMED Abstract]
- Carvajal RD, Antonescu CR, Wolchok JD, et al.: KIT as a therapeutic target in metastatic melanoma. JAMA 305 (22): 2327-34, 2011. [PUBMED Abstract]
- Gutzmer R, Stroyakovskiy D, Gogas H, et al.: Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAFV600 mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 395 (10240): 1835-1844, 2020. [PUBMED Abstract]
- Chapman PB, Einhorn LH, Meyers ML, et al.: Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with metastatic melanoma. J Clin Oncol 17 (9): 2745-51, 1999. [PUBMED Abstract]
- Middleton MR, Grob JJ, Aaronson N, et al.: Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol 18 (1): 158-66, 2000. [PUBMED Abstract]
- Avril MF, Aamdal S, Grob JJ, et al.: Fotemustine compared with dacarbazine in patients with disseminated malignant melanoma: a phase III study. J Clin Oncol 22 (6): 1118-25, 2004. [PUBMED Abstract]
- Robert C, Thomas L, Bondarenko I, et al.: Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 364 (26): 2517-26, 2011. [PUBMED Abstract]
- Ollila DW, Hsueh EC, Stern SL, et al.: Metastasectomy for recurrent stage IV melanoma. J Surg Oncol 71 (4): 209-13, 1999. [PUBMED Abstract]
- Gutman H, Hess KR, Kokotsakis JA, et al.: Surgery for abdominal metastases of cutaneous melanoma. World J Surg 25 (6): 750-8, 2001. [PUBMED Abstract]
Treatment of Stage 0 Melanoma
Treatment Options for Stage 0 Melanoma
Treatment options for stage 0 melanoma include:
Excision
There is no high-level evidence to guide the recommended excision margins for stage 0 (or in situ) melanoma. Consensus guidelines recommend margins of at least 5 mm for stage 0 melanoma, with a goal of achieving microscopically negative margins. However, 5 mm margins may be inadequate for some cases of in situ melanoma, and wider margins may be required.[1,2]
Current Clinical Trials
Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria. General information about clinical trials is also available.
References
- Kunishige JH, Doan L, Brodland DG, et al.: Comparison of surgical margins for lentigo maligna versus melanoma in situ. J Am Acad Dermatol 81 (1): 204-212, 2019. [PUBMED Abstract]
- Ellison PM, Zitelli JA, Brodland DG: Mohs micrographic surgery for melanoma: A prospective multicenter study. J Am Acad Dermatol 81 (3): 767-774, 2019. [PUBMED Abstract]
Treatment of Stage IA Melanoma
Treatment Options for Stage IA Melanoma
Treatment options for stage IA (pT1a or pT1b) melanoma include:
- Excision with or without sentinel lymph node biopsy (SLNB).
Excision
No randomized controlled trials have assessed only melanomas that are less than 1 mm thick. Evidence from randomized controlled clinical trials that included patients with melanomas of this size suggests that they may be adequately treated with radial excision margins of 1 cm.
Evidence (excision):
- A randomized trial that compared narrow margins (1 cm) with wide margins (≥3 cm) in 612 patients with melanomas no thicker than 2 mm included 359 patients with melanomas measuring 1 mm or less.[1,2][Level of evidence A1]
- No difference was observed between the two groups in the development of metastatic disease, disease-free survival (DFS), or overall survival (OS).
- There were no local recurrences in patients with melanomas measuring 1 mm or less in either cohort.
- A Swedish multicenter study included 989 patients with primary melanomas located on the trunk or extremities with a tumor thickness between 0.8 mm and 2 mm. Patients were randomly assigned to undergo wide excision with margins of 2 cm (n = 476) or 5 cm (n = 513).[3][Level of evidence A1]
- With a median follow-up of 11 years, there was no statistically significant difference in OS between the two groups.
- With a median follow-up of 8 years, there was no statistically significant difference in recurrence-free survival between the two groups.
- The local recurrence rate was 1%, and there was no significant difference between the treatment arms.
- A European, multicenter, randomized trial compared margins of 2 cm (n = 161) versus 5 cm (n = 165) in 326 patients with primary melanomas with a thickness of 2.1 mm or less. The study included 141 patients with melanomas measuring 1 mm or less. [3,4]Level of evidence A1]
- There was no statistically significant difference in 10-year DFS or OS between the two groups.
- Local recurrence occurred in one patient treated with a 2-cm margin and four patients treated with 5-cm margins.
Sentinel lymph node biopsy
Lymphatic mapping and SLNB for patients with high-risk, thin melanomas (≥0.8 mm) or ulcerated lesions measuring less than 0.8 mm may identify individuals with occult nodal disease. These procedures should be considered, particularly if other adverse prognostic features are present. Patients with clinically occult regional nodal metastases may benefit from ultrasound surveillance of regional lymph nodes and adjuvant therapy.[5-10]
Current Clinical Trials
Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria. General information about clinical trials is also available.
References
- Veronesi U, Cascinelli N: Narrow excision (1-cm margin). A safe procedure for thin cutaneous melanoma. Arch Surg 126 (4): 438-41, 1991. [PUBMED Abstract]
- Veronesi U, Cascinelli N, Adamus J, et al.: Thin stage I primary cutaneous malignant melanoma. Comparison of excision with margins of 1 or 3 cm. N Engl J Med 318 (18): 1159-62, 1988. [PUBMED Abstract]
- Cohn-Cedermark G, Rutqvist LE, Andersson R, et al.: Long term results of a randomized study by the Swedish Melanoma Study Group on 2-cm versus 5-cm resection margins for patients with cutaneous melanoma with a tumor thickness of 0.8-2.0 mm. Cancer 89 (7): 1495-501, 2000. [PUBMED Abstract]
- Balch CM, Soong SJ, Smith T, et al.: Long-term results of a prospective surgical trial comparing 2 cm vs. 4 cm excision margins for 740 patients with 1-4 mm melanomas. Ann Surg Oncol 8 (2): 101-8, 2001. [PUBMED Abstract]
- Essner R, Conforti A, Kelley MC, et al.: Efficacy of lymphatic mapping, sentinel lymphadenectomy, and selective complete lymph node dissection as a therapeutic procedure for early-stage melanoma. Ann Surg Oncol 6 (5): 442-9, 1999 Jul-Aug. [PUBMED Abstract]
- Gershenwald JE, Thompson W, Mansfield PF, et al.: Multi-institutional melanoma lymphatic mapping experience: the prognostic value of sentinel lymph node status in 612 stage I or II melanoma patients. J Clin Oncol 17 (3): 976-83, 1999. [PUBMED Abstract]
- Mraz-Gernhard S, Sagebiel RW, Kashani-Sabet M, et al.: Prediction of sentinel lymph node micrometastasis by histological features in primary cutaneous malignant melanoma. Arch Dermatol 134 (8): 983-7, 1998. [PUBMED Abstract]
- Morton DL, Thompson JF, Cochran AJ, et al.: Sentinel-node biopsy or nodal observation in melanoma. N Engl J Med 355 (13): 1307-17, 2006. [PUBMED Abstract]
- Faries MB, Thompson JF, Cochran AJ, et al.: Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N Engl J Med 376 (23): 2211-2222, 2017. [PUBMED Abstract]
- Leiter U, Stadler R, Mauch C, et al.: Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol 17 (6): 757-767, 2016. [PUBMED Abstract]
Treatment of Stage IB Melanoma
Treatment Options for Stage IB Melanoma
Treatment options for stage IB (pT2a) melanoma include:
Excision
No randomized controlled trials have compared 1-cm margins with 2-cm margins for melanomas measuring 2 mm or thinner. Evidence suggests that lesions no thicker than 2 mm may be treated conservatively with clinical radial excision margins of 1 cm to 2 cm. The decision to pursue a 1-cm versus a 2-cm margin should be based on patient and lesion factors, including functional and cosmetic limitations.
Evidence (excision):
- A randomized trial compared narrow margins (1 cm) with wide margins (≥3 cm) in 612 patients with melanomas no thicker than 2 mm.[1,2][Level of evidence A1]
- No difference was observed between the two groups in the development of metastatic disease, disease-free survival (DFS), or overall survival (OS).
- The risk of recurrence was 2.7% in patients with melanomas thicker than 1 mm who underwent narrow margins excision. No local recurrences occurred in the wider margin cohort.
- A Swedish multicenter study included 989 patients with primary melanomas located on the trunk or extremities with a tumor thickness between 0.8 mm and 2 mm. Patients were randomly assigned to undergo wide excision with margins of either 2 cm (n = 476) or 5 cm (n = 513).[3][Level of evidence A1]
- With a median follow-up of 11 years, there was no statistically significant difference in OS between the two treatment groups.
- With a median follow-up of 8 years, there was no statistically significant difference in recurrence-free survival between the two groups.
- The local recurrence rate was 1%, and there was no significant difference between the treatment arms.
- A European, multicenter, randomized trial compared margins of 2 cm (n = 161) versus 5 cm (n = 165) in 326 patients with primary melanomas with a thickness of 2.1 mm or less. The study included 141 patients with melanomas measuring 1 mm or less.[4][Level of evidence A1]
- There was no statistically significant difference in 10-year DFS or OS between the two treatment groups.
- Local recurrence occurred in one patient treated with a 2-cm margin and four patients treated with 5-cm margins.
- The Intergroup Melanoma Surgical Trial compared radial excision margins of 2 cm versus 4 cm in patients with melanomas of 1 mm to 4 mm in thickness.[5][Level of evidence A1]
- With a median follow-up of 10 years, there was no significant difference in OS, disease-specific survival, or local recurrence between the two treatment groups.
- A single-center retrospective study included 2,131 patients with melanomas between 1 mm and 2 mm (pT2) in thickness. The study compared excision with histological margins of 8 mm versus 16 mm (corresponding to 1-cm versus 2-cm clinical margins).[6][Level of evidence C1]
- There was no significant difference in 5-year melanoma-specific survival (P = .210) or 5-year DFS (P = .202) between the two groups.
- On multivariate analysis, peripheral excision margins did not influence local or in-transit recurrence.
- Another single-center retrospective study included 576 patients with melanomas of 1 mm to 2 mm (pT2) in thickness. The study compared excision with clinical margins of 1 cm versus 2 cm .[7][Level of evidence C2]
- The local recurrence rate was significantly higher in the 1-cm margin group than in the 2-cm margin group (3.6% vs. 0.9%; P = .044) on univariate analysis, but the rate was no longer significantly different on multivariate analysis.
- There was no difference in OS between the two groups.
Lymph node management
Elective regional lymph node dissection has no proven benefit for patients with stage I melanoma.[8]
Lymphatic mapping and sentinel lymph node biopsy (SLNB) for patients who have tumors of intermediate thickness may identify individuals with occult nodal disease. Patients with clinically occult regional nodal metastases may benefit from ultrasound surveillance of regional lymph nodes and adjuvant therapy.[9-14]
Evidence (SLNB versus observation):
- The International Multicenter Selective Lymphadenectomy Trial (MSLT-1) included 1,269 patients with intermediate-thickness (defined as 1.2–3.5 mm in this study) primary melanomas. The primary end point was melanoma-specific survival.[15][Level of evidence A1]
- At a median follow-up of 59.8 months, there was no melanoma-specific survival advantage for patients randomly assigned to undergo wide excision plus SLNB, followed by immediate completion lymphadenectomy for node positivity versus nodal observation and delayed lymphadenectomy for subsequent nodal recurrence.
- This trial was not designed to detect a difference in the impact of lymphadenectomy in patients with microscopic lymph node involvement.
Evidence (completion lymphadenectomy vs. observation with serial ultrasound of draining nodal basin):
- The Multicenter Selective Lymphadenectomy Trial II (MSLT-II [NCT00297895]) included 1,934 patients with primary melanomas and a positive sentinel node(s). Patients were randomly assigned to undergo either completion lymphadenectomy (n = 967) or nodal observation with ultrasound (n = 967). The primary end point was melanoma-specific survival.[13][Level of evidence A1]
- At a follow-up of 3 years, there was no melanoma-specific survival advantage for patients randomly assigned to undergo either completion lymphadenectomy versus nodal observation with ultrasound and delayed lymphadenectomy for subsequent nodal recurrence (86% [±1.3%] vs. 86% [±1.2%]; hazard ratio [HR], 1.08; 95% confidence interval [CI], 0.88–1.34; P = .42).
- At 3 years, the rate of nodal recurrence was 69% lower in the dissection group than in the observation group, with regional nodal disease control rates of 92% (±1.0%) versus 77% (±1.5%), respectively (HR, 0.31; 95% CI, 0.24–0.41; P < .001).
- With a median follow-up of 43 months, there was no significant difference in distant metastasis–free survival (DMFS) between the two groups (HR, 1.10; 95% CI, 0.92–1.31; P = .31).
- Adverse events were more common among the surgical cohort. Specifically, lymphedema occurred in 24.1% of patients in the dissection group versus 6.3% of patients in the observation group (P < .0001).
- A subgroup analysis did not identify any group (i.e., patients with >1 positive sentinel lymph node or a sentinel lymph node tumor >1 mm in diameter) that would benefit from a complete lymph node dissection.
- The DeCOG-SLT trial (NCT02434107) included 483 patients with primary melanoma and microscopically detected nodal metastases. Patients were randomly assigned to undergo either completion lymphadenectomy (n = 240) or observation (n = 233). The primary end point was DMFS.[14][Level of evidence B1]
- At a median follow-up of 35.5 months, there was no difference in DMFS in patients randomly assigned to undergo observation (77%; 90% CI, 71.9%–82.1%) versus completion lymphadenectomy (74.9%; 90% CI, 69.5%–80.3%) (HR, 1.03; 90% CI 0.71-1.50, P = 0.87).
- At 3 years, the OS rates were similar between patients randomly assigned to undergo observation (81.7%; 90% CI, 76.8%–86.6%) versus completion lymphadenectomy (81.2%; 90% CI, 76.1%–86.3%) (HR, 0.96; 90% CI, 0.67–1.38; P = .87), although the trial was closed early due to low event rates.
- In an exploratory analysis of DMFS in patients with micrometastasis thicker than 1 mm, there was no difference between the treatment groups (HR, 1.03; 90% CI, 0.63–1.71).
Current Clinical Trials
Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria. General information about clinical trials is also available.
References
- Veronesi U, Cascinelli N: Narrow excision (1-cm margin). A safe procedure for thin cutaneous melanoma. Arch Surg 126 (4): 438-41, 1991. [PUBMED Abstract]
- Veronesi U, Cascinelli N, Adamus J, et al.: Thin stage I primary cutaneous malignant melanoma. Comparison of excision with margins of 1 or 3 cm. N Engl J Med 318 (18): 1159-62, 1988. [PUBMED Abstract]
- Cohn-Cedermark G, Rutqvist LE, Andersson R, et al.: Long term results of a randomized study by the Swedish Melanoma Study Group on 2-cm versus 5-cm resection margins for patients with cutaneous melanoma with a tumor thickness of 0.8-2.0 mm. Cancer 89 (7): 1495-501, 2000. [PUBMED Abstract]
- Khayat D, Rixe O, Martin G, et al.: Surgical margins in cutaneous melanoma (2 cm versus 5 cm for lesions measuring less than 2.1-mm thick). Cancer 97 (8): 1941-6, 2003. [PUBMED Abstract]
- Balch CM, Urist MM, Karakousis CP, et al.: Efficacy of 2-cm surgical margins for intermediate-thickness melanomas (1 to 4 mm). Results of a multi-institutional randomized surgical trial. Ann Surg 218 (3): 262-7; discussion 267-9, 1993. [PUBMED Abstract]
- Haydu LE, Stollman JT, Scolyer RA, et al.: Minimum Safe Pathologic Excision Margins for Primary Cutaneous Melanomas (1-2 mm in Thickness): Analysis of 2131 Patients Treated at a Single Center. Ann Surg Oncol 23 (4): 1071-81, 2016. [PUBMED Abstract]
- Hudson LE, Maithel SK, Carlson GW, et al.: 1 or 2 cm margins of excision for T2 melanomas: do they impact recurrence or survival? Ann Surg Oncol 20 (1): 346-51, 2013. [PUBMED Abstract]
- Hochwald SN, Coit DG: Role of elective lymph node dissection in melanoma. Semin Surg Oncol 14 (4): 276-82, 1998. [PUBMED Abstract]
- Essner R, Conforti A, Kelley MC, et al.: Efficacy of lymphatic mapping, sentinel lymphadenectomy, and selective complete lymph node dissection as a therapeutic procedure for early-stage melanoma. Ann Surg Oncol 6 (5): 442-9, 1999 Jul-Aug. [PUBMED Abstract]
- Gershenwald JE, Thompson W, Mansfield PF, et al.: Multi-institutional melanoma lymphatic mapping experience: the prognostic value of sentinel lymph node status in 612 stage I or II melanoma patients. J Clin Oncol 17 (3): 976-83, 1999. [PUBMED Abstract]
- Mraz-Gernhard S, Sagebiel RW, Kashani-Sabet M, et al.: Prediction of sentinel lymph node micrometastasis by histological features in primary cutaneous malignant melanoma. Arch Dermatol 134 (8): 983-7, 1998. [PUBMED Abstract]
- Morton DL, Thompson JF, Cochran AJ, et al.: Sentinel-node biopsy or nodal observation in melanoma. N Engl J Med 355 (13): 1307-17, 2006. [PUBMED Abstract]
- Faries MB, Thompson JF, Cochran AJ, et al.: Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N Engl J Med 376 (23): 2211-2222, 2017. [PUBMED Abstract]
- Leiter U, Stadler R, Mauch C, et al.: Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol 17 (6): 757-767, 2016. [PUBMED Abstract]
- Morton DL, Thompson JF, Cochran AJ, et al.: Final trial report of sentinel-node biopsy versus nodal observation in melanoma. N Engl J Med 370 (7): 599-609, 2014. [PUBMED Abstract]
Treatment of Stage II Melanoma
Treatment Options for Stage II Melanoma
Treatment options for stage II melanoma include:
Excision
Evidence suggests that lesions no thicker than 2 mm (pT2b) may be treated conservatively with clinical radial excision margins of 1 cm to 2 cm. The decision to pursue a 1-cm versus 2-cm margin should be based on patient and lesion factors, including functional and cosmetic limitations. Evidence suggests that for melanomas measuring 2 mm or thicker (pT3a/b, pT4a/b), clinical radial margins of 2 cm are recommended. There is no evidence to support that a wider margin is beneficial.[1]
Evidence (excision):
- The Intergroup Melanoma Surgical Trial Task 2b compared 2-cm versus 4-cm margins for 740 patients with melanomas that were 1 mm to 4 mm thick.[2]
- With a median follow-up of more than 10 years, no significant difference in local recurrence or survival was observed between the two groups.
- The reduction in margins from 4 cm to 2 cm was associated with a statistically significant reduction in the need for skin grafting (from 46% to 11%; P < .001).
- A study conducted in the United Kingdom randomly assigned patients with melanomas thicker than 2 mm to undergo excision with either 1-cm or 3-cm margins.[3,4][Level of evidence A1]
- At a median follow-up of 5 years, patients who underwent excision with 1-cm margins had higher rates of locoregional recurrence (hazard ratio [HR], 1.34; 95% confidence interval [CI], 1.06–1.71; P = .02).
- No difference in overall survival (OS) was seen (HR, 0.81; 95% CI, 0.58–1.13; P = .386) between the two groups. However, at 8.8 years, there was a melanoma-specific survival advantage for patients who underwent excision with 3-cm margins compared with 1-cm margins (HR, 1.24; 95% CI, 1.01–1.52; P = .036).
- This study suggests that 1-cm margins may not be adequate for patients with melanomas thicker than 2 mm.
- In a multicenter international trial (NCT03638492), the Swedish and Danish melanoma groups randomly assigned patients with localized cutaneous melanoma thicker than 2mm to undergo excision with clinical margins of either 2 cm (n = 471) or 4 cm (n = 465).[1][Level of evidence A1]
- With a median follow-up of 19.6 years, no significant difference in OS or melanoma-specific survival was observed between the two groups (HR, 0.98; 95% CI, 0.83–1.14; P = .75).
Lymph node management
Lymphatic mapping and sentinel lymph node biopsy (SLNB)
Lymphatic mapping and SLNB have assessed the presence of occult metastasis in the regional lymph nodes of patients with stage II disease. These procedures may identify individuals who can avoid regional lymph node dissection and individuals who may benefit from adjuvant therapy.[5-9]
To ensure accurate identification of the sentinel lymph node, lymphatic mapping and removal of the sentinel lymph node are performed during the same operation as the wide excision of the primary melanoma.
With the use of a vital blue dye and a radiopharmaceutical agent injected at the site of the primary tumor, the first lymph node in the lymphatic basin that drains the lesion can be identified, removed, and examined microscopically. Multiple studies have demonstrated the diagnostic accuracy of SLNB, with false-negative rates of 0% to 2%.[5,10-14]
Regional lymphadenectomy
In patients with microscopic melanoma in regional lymph nodes, immediate completion lymphadenectomy has widely been replaced by active observation, as long as close follow-up with nodal ultrasound surveillance can be achieved.[15,16] Completion lymphadenectomy may still be considered on a case-by-case basis.
Evidence (completion lymphadenectomy vs. observation with serial ultrasound of draining nodal basin):
- The Multicenter Selective Lymphadenectomy Trial II (MSLT-II [NCT00297895]) included 1,934 patients with primary melanomas and a positive sentinel node(s). Patients were randomly assigned to undergo either completion lymphadenectomy (n = 967) or nodal observation with ultrasound (n = 967). The primary end point was melanoma-specific survival.[15][Level of evidence A1]
- After a follow-up of 3 years, there was no melanoma-specific survival advantage for patients randomly assigned to undergo completion lymphadenectomy versus nodal observation with ultrasound and delayed lymphadenectomy for subsequent nodal recurrence (86% [±1.3%] vs. 86% [±1.2%]; HR, 1.08; 95% CI, 0.88–1.34; P = .42).
- At 3 years, the rate of nodal recurrence was 69% lower in the dissection group than in the observation group, with regional nodal disease control rates of 92% (±1.0%) versus 77% (±1.5%), respectively (HR, 0.31; 95% CI, 0.24–0.41; P < .001).
- With a median follow-up of 43 months, there was no significant difference in distant metastasis–free survival (DMFS) between the two groups (HR, 1.10; 95% CI, 0.92–1.31; P = .31).
- Adverse events were more common among the surgical cohort. Specifically, lymphedema occurred in 24.1% of patients in the dissection group versus 6.3% of patients in the observation group (P < .0001).
- A subgroup analysis did not identify any group (i.e., patients with >1 positive sentinel lymph node or a sentinel lymph node tumor diameter >1 mm) that would benefit from a complete lymph node dissection.
- The DeCOG-SLT trial (NCT02434107) included 483 patients with primary melanoma and microscopically detected nodal metastases. Patients were randomly assigned to undergo completion lymphadenectomy (n = 240) or observation (n = 233). The primary end point was DMFS.[16][Level of evidence B1]
- At a median follow-up of 35.5 months, there was no difference in DMFS between patients randomly assigned to undergo observation (77%; 90% CI, 71.9%–82.1%) versus completion lymphadenectomy (74.9%; 90% CI, 69.5%–80.3%) (HR, 1.03; 90% CI, 0.71–1.50; P = .87).
- At 3 years, the OS rates were similar between patients randomly assigned to undergo observation (81.7%; 90% CI, 76.8%–86.6%) versus completion lymphadenectomy (81.2%; 90% CI, 76.1%–86.3%) (HR, 0.96; 90% CI, 0.67–1.38; P = .87), although the trial was closed early due to low event rates.
- In an exploratory analysis of DMFS in patients with micrometastasis thicker than 1 mm, there was no difference between the treatment groups (HR, 1.03; 90% CI, 0.63–1.71).
Adjuvant therapy
Adjuvant therapeutic options are expanding for patients at high risk of recurrence after complete resection. Patients with resected stage IIB or stage IIC melanoma, despite not having lymph node involvement, have a similar risk of recurrence and melanoma-specific death as patients with stage III melanoma. As outlined in the Treatment of Resectable Stage III Melanoma section, the U.S. Food and Drug Administration (FDA) has approved several agents as adjuvant therapy for patients with resected stage III melanoma.
Immunotherapy
Adjuvant immunotherapy has demonstrated a recurrence-free survival (RFS) and DMFS benefit in patients with high-risk stage II melanoma that has been resected. These results led to FDA approval for the anti–programmed death-1 agent pembrolizumab. Data regarding potential OS benefit are still pending.
Checkpoint inhibitors
Pembrolizumab
Evidence (pembrolizumab):
- In a multinational double-blind trial (KEYNOTE-716 [NCT03553836]), patients with completely resected stage IIB or stage IIC melanoma were randomly assigned (1:1) to receive either pembrolizumab or placebo. The primary end point was RFS, defined as the time from randomization until the date of first recurrence or death from any cause. If recurrence was documented, patients could cross over or repeat treatment with pembrolizumab. The secondary end point was DMFS. Pembrolizumab was given as an intravenous infusion of 200 mg every 3 weeks, for a total of 17 doses (approximately 1 year).
A total of 976 patients were randomly assigned: 487 to pembrolizumab and 489 to placebo. Baseline characteristics were balanced, including the number of patients with ulcerated tumors (40% of stage IIB; 36% of stage IIC) in both arms.[17]
- At the first interim analysis with a median follow-up of 14.4 months, the 12-month RFS rate was 90.0% (95% CI, 87%–93%) in the pembrolizumab group and 83% (95% CI, 79%–86%) in the placebo group.[17][Level of evidence B1]
- At the second interim analysis with a median follow-up of 20.9 months, the 18-month RFS rate was 86% (95% CI, 82%–89%) in the pembrolizumab group and 77% (95% CI, 73%–81%) in the placebo group.
- At a median follow-up of 39.4 months, the 36-month RFS rate was 76.2% in the pembrolizumab group and 63.4% in the placebo group. An updated analysis of DMFS reported a 36-month DMFS rate of 84.4% in the pembrolizumab group and 74.7% in the placebo group. Median DMFS was not reached in either group.[18,19]
Current Clinical Trials
Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria. General information about clinical trials is also available.
References
- Utjés D, Malmstedt J, Teras J, et al.: 2-cm versus 4-cm surgical excision margins for primary cutaneous melanoma thicker than 2 mm: long-term follow-up of a multicentre, randomised trial. Lancet 394 (10197): 471-477, 2019. [PUBMED Abstract]
- Balch CM, Urist MM, Karakousis CP, et al.: Efficacy of 2-cm surgical margins for intermediate-thickness melanomas (1 to 4 mm). Results of a multi-institutional randomized surgical trial. Ann Surg 218 (3): 262-7; discussion 267-9, 1993. [PUBMED Abstract]
- Thomas JM, Newton-Bishop J, A'Hern R, et al.: Excision margins in high-risk malignant melanoma. N Engl J Med 350 (8): 757-66, 2004. [PUBMED Abstract]
- Hayes AJ, Maynard L, Coombes G, et al.: Wide versus narrow excision margins for high-risk, primary cutaneous melanomas: long-term follow-up of survival in a randomised trial. Lancet Oncol 17 (2): 184-192, 2016. [PUBMED Abstract]
- Gershenwald JE, Thompson W, Mansfield PF, et al.: Multi-institutional melanoma lymphatic mapping experience: the prognostic value of sentinel lymph node status in 612 stage I or II melanoma patients. J Clin Oncol 17 (3): 976-83, 1999. [PUBMED Abstract]
- McMasters KM, Reintgen DS, Ross MI, et al.: Sentinel lymph node biopsy for melanoma: controversy despite widespread agreement. J Clin Oncol 19 (11): 2851-5, 2001. [PUBMED Abstract]
- Cherpelis BS, Haddad F, Messina J, et al.: Sentinel lymph node micrometastasis and other histologic factors that predict outcome in patients with thicker melanomas. J Am Acad Dermatol 44 (5): 762-6, 2001. [PUBMED Abstract]
- Essner R: The role of lymphoscintigraphy and sentinel node mapping in assessing patient risk in melanoma. Semin Oncol 24 (1 Suppl 4): S8-10, 1997. [PUBMED Abstract]
- Chan AD, Morton DL: Sentinel node detection in malignant melanoma. Recent Results Cancer Res 157: 161-77, 2000. [PUBMED Abstract]
- Morton DL, Wen DR, Wong JH, et al.: Technical details of intraoperative lymphatic mapping for early stage melanoma. Arch Surg 127 (4): 392-9, 1992. [PUBMED Abstract]
- Reintgen D, Cruse CW, Wells K, et al.: The orderly progression of melanoma nodal metastases. Ann Surg 220 (6): 759-67, 1994. [PUBMED Abstract]
- Thompson JF, McCarthy WH, Bosch CM, et al.: Sentinel lymph node status as an indicator of the presence of metastatic melanoma in regional lymph nodes. Melanoma Res 5 (4): 255-60, 1995. [PUBMED Abstract]
- Uren RF, Howman-Giles R, Thompson JF, et al.: Lymphoscintigraphy to identify sentinel lymph nodes in patients with melanoma. Melanoma Res 4 (6): 395-9, 1994. [PUBMED Abstract]
- Bostick P, Essner R, Glass E, et al.: Comparison of blue dye and probe-assisted intraoperative lymphatic mapping in melanoma to identify sentinel nodes in 100 lymphatic basins. Arch Surg 134 (1): 43-9, 1999. [PUBMED Abstract]
- Faries MB, Thompson JF, Cochran AJ, et al.: Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N Engl J Med 376 (23): 2211-2222, 2017. [PUBMED Abstract]
- Leiter U, Stadler R, Mauch C, et al.: Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol 17 (6): 757-767, 2016. [PUBMED Abstract]
- Luke JJ, Rutkowski P, Queirolo P, et al.: Pembrolizumab versus placebo as adjuvant therapy in completely resected stage IIB or IIC melanoma (KEYNOTE-716): a randomised, double-blind, phase 3 trial. Lancet 399 (10336): 1718-1729, 2022. [PUBMED Abstract]
- Long GV, Luke JJ, Khattak MA, et al.: Pembrolizumab versus placebo as adjuvant therapy in resected stage IIB or IIC melanoma (KEYNOTE-716): distant metastasis-free survival results of a multicentre, double-blind, randomised, phase 3 trial. Lancet Oncol 23 (11): 1378-1388, 2022. [PUBMED Abstract]
- Luke JJ, Ascierto PA, Khattak MA, et al.: Pembrolizumab Versus Placebo as Adjuvant Therapy in Resected Stage IIB or IIC Melanoma: Final Analysis of Distant Metastasis-Free Survival in the Phase III KEYNOTE-716 Study. J Clin Oncol 42 (14): 1619-1624, 2024. [PUBMED Abstract]
Treatment of Resectable Stage III Melanoma
Treatment Options for Resectable Stage III Melanoma
Treatment options for resectable stage III melanoma include the following:
- Excision with or without lymph node management.
- Neoadjuvant therapy.
- Adjuvant therapy.
- Combination immunotherapies, including vaccines (under clinical evaluation).
- Adjuvant therapies that target a known pathogenic variant (e.g., KIT) (under clinical evaluation).
- Intralesional therapies (under clinical evaluation).
Excision
The primary tumor may be treated with wide local excision with 1-cm to 2-cm margins, depending on tumor thickness and location.[1-7] Skin grafting may be necessary to close the resulting defect.
Lymph node management
Sentinel lymph node biopsy (SLNB)
Lymphatic mapping and SLNB can be considered to assess the presence of occult metastases in the regional lymph nodes of patients with primary tumors measuring at least 0.8 mm thick. These procedures may identify individuals who can avoid regional lymph node dissection and individuals who may benefit from adjuvant therapy.[3,8-12]
To ensure accurate identification of the sentinel lymph node, lymphatic mapping and removal of the sentinel lymph node are performed during the same operation as the wide excision of the primary melanoma.
Multiple studies have demonstrated the diagnostic accuracy of SLNB, with false-negative rates of 0% to 2%.[8,12-17] If micrometastatic melanoma is detected, active surveillance with ultrasound of the draining nodal basin is an acceptable treatment recommendation that has widely replaced complete lymph node dissection (CLND).[18,19] A complete regional lymphadenectomy can be considered in select populations.
Regional lymphadenectomy
In patients with microscopic melanoma in regional lymph nodes, immediate completion lymphadenectomy has widely been replaced by active observation, as long as close follow-up with nodal ultrasound surveillance can be achieved. Completion lymphadenectomy may still be considered on a case-by-case basis.
For patients who present with clinically detected (i.e., macroscopic) regional lymph node involvement in the absence of distant disease, therapeutic regional lymphadenectomy may be performed.
Neoadjuvant therapy
Resection of the bulk of the tumor and tumor-infiltrating lymphocytes may decrease the potential effect of programmed death-1 (PD-1) blockade in the adjuvant setting, leading investigators to study the use of neoadjuvant immunotherapy. Neoadjuvant pembrolizumab can be considered for patients with high-risk, node-positive disease or resectable metastatic disease. This treatment approach is based on the results of a randomized phase II trial in which 1 year of neoadjuvant and adjuvant pembrolizumab resulted in an improvement in event-free survival (EFS) compared with primary resection plus adjuvant pembrolizumab. The U.S. Food and Drug Administration (FDA) has not approved this regimen, pending the completion of a randomized phase III trial.
Immunotherapy
Checkpoint inhibitors
Pembrolizumab
Evidence (pembrolizumab):
- An open-label phase II study (SWOG S1801 [NCT03698019]) conducted at 90 sites in the United States enrolled 313 patients with resectable macroscopic stage IIIB to stage IV melanoma. Patients were randomly assigned (1:1) to receive either three cycles of neoadjuvant pembrolizumab (200 mg intravenously [IV] every 3 weeks) followed by surgery and fifteen cycles of adjuvant pembrolizumab (n = 154 patients) or primary surgery followed by eighteen cycles of adjuvant pembrolizumab (n = 159 patients).[20] The primary end point was EFS in the intention-to-treat population. Events were defined as disease progression or toxic effects that precluded surgery; the inability to resect all gross disease; disease progression, surgical complications, or toxic effects of treatment that precluded the initiation of adjuvant therapy within 84 days after surgery; recurrence of melanoma after surgery; or death from any cause. Exclusion criteria included prior immunotherapy for melanoma, active autoimmune disease in patients who had received systemic treatment within 2 years before trial entry, uveal melanoma, and any history of brain metastasis.
- The median duration of follow-up was 14.7 months in both groups. A total of 105 events occurred (38 in the neoadjuvant-adjuvant group and 67 in the adjuvant-only group). EFS was significantly longer in the neoadjuvant-adjuvant group than in the adjuvant-only group (P = .004). The EFS rate at 2 years was 72% (95% confidence interval [CI], 64%–80%) in the neoadjuvant-adjuvant group and 49% (95% CI, 41%–59%) in the adjuvant-only group.[20][Level of evidence B1]
- At the time of data cutoff, 36 deaths (14 in the neoadjuvant-adjuvant group and 22 in the adjuvant-only group) were reported. This small number of deaths precluded a comparison of overall survival (OS). The benefit of neoadjuvant-adjuvant pembrolizumab was seen across all subgroups of patients including those with BRAF pathogenic variants.
- Among the 152 patients in the neoadjuvant–adjuvant group who had received at least one dose of pembrolizumab, 11 (7%) had at least one grade 3 or 4 adverse event that was deemed by the investigators to be related to pembrolizumab. The incidence of adverse events of grade 3 or higher during adjuvant therapy was similar in the two groups (12% in the neoadjuvant-adjuvant group and 14% in the adjuvant-only group). No new toxic effects of pembrolizumab were observed in either trial group. No deaths attributed by the investigators to pembrolizumab occurred in either group.
Ipilimumab and nivolumab
Evidence (ipilimumab and nivolumab):
- An open-label phase III study (NADINA [NCT04949113]) included patients with resectable macroscopic stage III melanoma. Patients were randomly assigned (1:1) to receive either two cycles of neoadjuvant ipilimumab plus nivolumab every 3 weeks, followed by lymph node dissection/resection of in-transit metastases or lymph node dissection followed by 12 cycles of adjuvant nivolumab every 4 weeks. A total of 423 eligible patients were randomly assigned (212 patients to the neoadjuvant group and 211 to the adjuvant-only group). Adjuvant therapy for patients assigned to the neoadjuvant group was dictated by pathological response. Patients who had a major pathological response (≤10% residual viable tumor) did not receive any adjuvant treatment. Patients who had a pathological partial response (11%–50%) or a pathological nonresponse (>50% residual tumor) received either adjuvant dabrafenib plus trametinib for 46 weeks or an additional 11 cycles of adjuvant
nivolumab. The primary end point was EFS in the intention-to-treat population. Events were defined as the time from randomization to the occurrence of progression to unresectable melanoma before surgery, disease recurrence, or death due to melanoma or treatment.[21]
- The median duration of follow-up was 10.6 months in both groups. A total of 100 events occurred (28 in the neoadjuvant-adjuvant group and 72 in the adjuvant-only group). EFS was significantly longer in the neoadjuvant group than in the adjuvant-only group. The 12-month EFS rate was 83.7% (95% CI, 73.8%–94.8%) in the neoadjuvant group and 57.2% (95% CI, 45.1%–72.7%) in the adjuvant-alone group.[21][Level of evidence B1] The benefit of neoadjuvant therapy was seen across all subgroups of patients, including those with BRAF pathogenic variants.
- Among the 212 patients in the neoadjuvant group who received at least one dose of treatment, 47.2% had at least one grade 3 or 4 adverse event, compared with 34.1% in the adjuvant-only group. No new toxic effects of ipilimumab and nivolumab were observed in either trial group. One patient in the adjuvant group died of pneumonitis caused by treatment. No treatment-related deaths occurred in the neoadjuvant group.
Adjuvant therapy
Adjuvant therapeutic options are expanding for patients at high risk of recurrence after complete resection and include checkpoint inhibitors and combination signal transduction inhibitor therapy. Ipilimumab was the first checkpoint inhibitor to be approved by the FDA as adjuvant therapy. It has demonstrated improved OS when given at 10 mg/kg (ipi10), compared with placebo (EORTC 18071 [NCT00636168]).[22] However, ipi10 has significant toxicity at this dose. The North American Intergroup Trial E1609 (NCT01274338), designed with three treatment groups, compared ipi10 with ipilimumab at a lower dose of 3 mg/kg (ipi3) (approved for metastatic disease) and with high-dose interferon (HDI). Patients who received ipi3 had a significant improvement in OS compared with ipi10.[23] These data do not support HDI as an adjuvant treatment option for melanoma. As newer checkpoint inhibitors emerge, the role of ipi3 remains to be further defined.
Large randomized trials with the newer checkpoint inhibitors (nivolumab and pembrolizumab) and with combination signal transduction inhibitors (dabrafenib plus trametinib) showed a clinically significant impact on relapse-free survival (RFS). The CheckMate 238 (NCT02388906) trial compared nivolumab with ipi10 and found that nivolumab produced superior RFS and had a more tolerable safety profile.[24] Pembrolizumab produced superior RFS compared with placebo, with data on OS still maturing in the MK-3475-054/KEYNOTE-054 trial (NCT02362594).[25] Dabrafenib plus trametinib produced superior RFS compared with placebo, with data on OS still maturing in the COMBI-AD trial (NCT01682083).[26] Single-agent BRAF-inhibitor therapy with vemurafenib did not show improved RFS compared with placebo in the BRIM8 trial (NCT01667419).[27]
The benefit of immunotherapy with ipilimumab, nivolumab, and pembrolizumab has been seen regardless of programmed death-ligand 1 (PD-L1) expression or the presence of BRAF pathogenic variants. Combination signal transduction inhibitor therapy with dabrafenib plus trametinib is an additional option for patients with BRAF variants.
Patients should consider participating in clinical trials to identify treatments that will further extend RFS and OS with less toxicity and shorter treatment schedules.
Immunotherapy
Checkpoint inhibitors
Nivolumab
Evidence (nivolumab):
- In a multinational double-blind trial (CheckMate 238 [NCT02388906]), patients with stage IIIB, IIIC, or IV melanoma who underwent complete resection were randomly assigned (1:1) to receive either nivolumab or ipilimumab.[24][Level of evidence B1] The primary end point was RFS and was defined as time from randomization until the date of the first recurrence, new primary melanoma, or death from any cause. Patients who were excluded included those with resection occurring more than 12 weeks before randomization, autoimmune disease, use of systemic glucocorticoids, previous systemic therapy for melanoma, and an Eastern Cooperative Oncology Group (ECOG) Performance Status (PS) score higher than 1. Nivolumab (3 mg/kg) was given IV every 2 weeks and ipilimumab (10 mg/kg) was given every 3 weeks for four doses, then every 3 months for up to 1 year or
until disease recurrence, along with corresponding placebo.
A total of 906 patients were randomly assigned: 453 patients to nivolumab and 453 patients to ipilimumab. Baseline characteristics were balanced in the two treatment groups. Approximately 81% of patients had stage III disease, 32% had ulcerated primary melanoma, 48% had macroscopic lymph node involvement, 62% had less than a 5% PD-L1 expression, and 42% had BRAF pathogenic variants.
- The European Organisation for Research and Treatment of Cancer (EORTC) Independent Data Monitoring Committee stopped the study at the protocol-specified interim analysis, when all patients had a minimum follow-up of 18 months, at which time there were 360 events of RFS. The median RFS has not been reached in either treatment group. At 12 months, the RFS rate was 70.5% (95% CI, 66.1%–74.0%) for patients treated with nivolumab versus 60.8% (95% CI, 56.0%–65.2%) for patients treated with ipilimumab. Recurrence or death occurred in 34% (154 of 453) of patients treated with nivolumab versus 45.5% (206 of 453) of patients treated with ipilimumab (hazard ratio [HR] recurrence or death, 0.65; 97.56% CI, 0.51–0.83; P < .001). Subgroup analyses of RFS favored nivolumab regardless of PD-L1 expression or the presence of a BRAF V600 pathogenic variant.
- Patients treated with nivolumab had fewer adverse events, including grade 3 to 4 serious adverse events and death. Adverse events led to treatment discontinuation in 9.7% of patients assigned to nivolumab and 42.6% of patients assigned to ipilimumab. Two treatment-related deaths occurred among patients treated with ipilimumab (e.g., marrow aplasia and colitis) and none occurred among the patients treated with nivolumab. The adverse event profile was like the type of checkpoint inhibitor toxicities seen in the metastatic setting, with immune-related events most commonly seen in the gastrointestinal system, hepatic system, and skin. Grade 3 or 4 adverse events occurred in 14% of patients treated with nivolumab and in 46% of patients treated with ipilimumab.
- An updated analysis, after a minimum of 4 years of follow-up, showed an RFS rate of 51.7% (95% CI, 46.8%–56.3%) for patients who received nivolumab versus 41.2% (95% CI, 36.4%–45.9%) for patients who received ipilimumab (HR relapse or death, 0.71; 95% CI, 0.60–0.86; P = .0003). Median OS has not been reached in either group.[28]
Pembrolizumab
Evidence (pembrolizumab):
- In a multinational double-blind trial (MK-3475-054/KEYNOTE-054 [NCT02362594]), patients with completely resected stage IIIA, IIIB, or IIIC melanoma were randomly assigned (1:1) to receive either pembrolizumab or placebo.[25][Level of evidence B1] The primary end point was RFS, defined as time from randomization until the date of first recurrence or death from any cause. If recurrence was documented, patients could cross over or repeat treatment with pembrolizumab. Pembrolizumab was given as an IV infusion of 200 mg every 3 weeks, for a total of 18 doses (approximately 1 year).
A total of 1,019 patients were randomly assigned: 514 to pembrolizumab and 505 to placebo. Baseline characteristics were balanced in the two treatment groups. Approximately 40% had ulcerated primary melanoma, 66% had macroscopic lymph node involvement, 84% had positive PD-L1 expression (melanoma score >2 by 22C3 antibody assay), and 44% had BRAF pathogenic variants.
- The EORTC Independent Data Monitoring Committee reviewed the unblinded results at an amended interim analysis when 351 events (recurrences or deaths) had occurred. The results were positive, and the interim analysis of RFS became the final analysis.
- At the amended interim analysis with a median follow-up of 15 months, the 12-month RFS rate was 75.4% (95% CI, 71.3%–78.9%) in the pembrolizumab group versus 61.0% (95% CI, 56.5%–65.1%) in the placebo group.
- An update of the primary end point of RFS with a median follow-up 3.5 years showed an RFS rate of 59.8% (95% CI, 55.3%–64.1%) in the pembrolizumab group versus 41.4% (95% CI, 39.2%–48.8%) in the placebo group (HR, 0.59; 95% CI, 0.49–0.73).[29]
- Pembrolizumab maintained its effect regardless of PD-L1 expression or BRAF pathogenic variant status.
- Approximately 14% of patients discontinued pembrolizumab due to an adverse event. Grade 3, 4, or 5 adverse events considered to be related to pembrolizumab occurred in 15% of patients. There was one death due to treatment (myositis).
Ipilimumab
Evidence (ipilimumab):
- The open-label, three-arm, North American Intergroup trial E1609 (NCT01274338) compared two doses of ipilimumab with HDI as adjuvant therapy in high-risk patients with melanoma.[23] A total of 1,670 patients with resected disease (defined by the American Joint Committee on Cancer, 7th edition, as stage IIIB, IIIC, M1a, or M1b) were randomly assigned (1:1:1) to ipi3 or ipi10 every 3 weeks for four doses (induction), followed by the same dose every 12 weeks for four doses (maintenance), or HDI 20 million units/m2 per day, 5 days per week for 4 weeks (induction), followed by 10 million units/m2 daily subcutaneously every other day, 3 days per week for 48 weeks (maintenance).[23][Level of evidence A1]
The trial was designed with two coprimary end points, RFS and OS, with a hierarchic analysis to evaluate ipi3 versus HDI followed by ipi10 versus HDI. The time to event was longer than anticipated and the design was amended for a final analysis at a data cutoff date giving a median follow-up time of 57.4 months (range, 0.03−86.6).
- Ipi3 significantly improved OS compared with HDI (HR, 0.78; 95% repeated CI, 0.61−0.99; P = .044), but not RFS (HR, 0.85; 95% CI, 0.66−1.09; P = .065).
- Ipi10 did not significantly improve OS or RFS compared with HDI (HR, 0.88; 95.6% CI, 0.69−1.12 and exploratory HR, 0.84; 99.4% CI, 0.65−1.09, respectively).
- Salvage treatments were used in 69.7% of patients after ipi3, 51.6% of patients after ipi10, and 86.2% of patients after HDI.
Toxicity with ipi3 was lower than with ipi10; however, both had treatment-related discontinuations and death.
- Treatment-related discontinuations occurred in 34.9% of the ipi3 group and in 54.1% of the ipi10 group.
- Three possibly treatment-related deaths occurred in the ipi3 group, five in the ipi10 group, and two in the HDI group.
The study concluded that evidence no longer supports a role for HDI as adjuvant therapy for patients with high-risk melanoma. Further, ipi3 provides OS data superior to ipi10 compared with HDI. The role of ipilimumab as adjuvant monotherapy is unclear because the CheckMate 238 trial demonstrated that nivolumab was superior to ipi10 in improving RFS, with OS data still maturing.
- In a multinational double-blind trial (EORTC 18071 [NCT00636168]), patients with stage III melanoma, who had complete resection, were randomly assigned (1:1) to receive either ipilimumab or placebo.[30][Level of evidence B1] Exclusion criteria comprised patients with lymph node metastasis larger than 1 mm, in-transit metastasis, resection occurring more than 12 weeks before randomization, autoimmune disease, previous or concurrent immunosuppressive therapy, previous systemic therapy for melanoma, and an ECOG PS score higher than 1. The ipilimumab dose was 10 mg/kg every 3 weeks for four doses, then every 3 months for up to 3 years. The primary end point was RFS, defined as recurrence or death (regardless of cause), whichever came first, as assessed by an independent review committee.
- A total of 951 patients were enrolled (475 patients to the ipilimumab arm and 476 patients to the placebo arm). The median age was 51 years, and 94% of the patients had a PS of 0.
- At a median follow-up of 2.7 years, there were 528 RFS events: 234 in the ipilimumab group (49%; 220 recurrences, 14 deaths) and 294 in the placebo group (62%; 289 recurrences, 5 deaths). Median RFS was 26 months for the ipilimumab group (95% CI, 19–39) versus 17 months for the placebo group (95% CI, 13–22). The HR was 0.75 (95% CI, 0.64–0.90; P < .002). The effect of ipilimumab was consistent across subgroups.
- Ipilimumab was discontinued because of adverse events in 52% of the patients. Patients received ipilimumab for a median of four doses; 36% of patients in the ipilimumab group stayed on treatment for more than 6 months, and 26% stayed on treatment for more than 1 year. Five patients died from drug-related events: three secondary to colitis, one with myocarditis, and one of multiorgan failure with Guillain-Barré syndrome. The most common adverse events were gastrointestinal, hepatic, and endocrine related and included rash, fatigue, and headache.
An updated analysis was performed at a median follow-up of 5.3 years.[22]
- The 5-year RFS rate was 40.8% in patients treated with ipilimumab and 30.3% in patients who received the placebo (HR recurrence or death, 0.76; 95% CI, 0.64–0.89; P < .001). The median RFS rate was 27.6% in patients treated with ipilimumab and 17.1% in patients who received the placebo.
- The 5-year OS rate, a secondary end point, was 65.4% in patients treated with ipilimumab versus 54.4% in patients who received the placebo (HRdeath, 0.72; 95.1% CI, 0.58–0.88; P = .001).
Data from this trial (EORTC 18071), which tested high-dose ipilimumab at 10 mg/kg compared with placebo, served as the basis for the approval of ipilimumab in the adjuvant setting. However, the subsequent intergroup trial, E1609 (described above), demonstrated better outcomes with low-dose (3 mg/kg) ipilimumab, which is also the dose approved for metastatic disease.
Combination signal transduction inhibitors
Dabrafenib plus trametinib
Evidence (dabrafenib plus trametinib):
- A multinational double-blind trial (COMBI-AD [NCT01682083]) included patients with stage IIIA, IIIB, or IIIC melanoma with BRAF V600E or V600K pathogenic variants who underwent completion lymphadenectomy. Patients were randomly assigned (1:1) to receive either dabrafenib plus trametinib or two matched placebo tablets.[26] The primary end point was RFS, defined as time from randomization until the date of first recurrence or death from any cause. Patients with resection occurring more than 12 weeks before random assignment and an ECOG PS score higher than 1 were excluded. Dabrafenib was given at a dose of 150 mg twice daily plus trametinib at a dose of 2 mg once daily (combination therapy) for 12 months in the absence of disease recurrence, unacceptable toxic effects, or death. A total of 870 patients were randomly assigned (438 patients to combination therapy and 432 patients to placebo). Baseline characteristics were balanced in the two treatment groups. Most patients (91%) had BRAF V600E pathogenic variants compared with 9% who had BRAF V600K variants. Most patients (92%) had an ECOG PS of 0.
- At the data cutoff date for the primary analysis, the minimum follow-up was 2.5 years (median, 2.8 years), and all patients had completed trial treatment. Disease recurrence occurred in 163 of 438 patients (37%) who received combination therapy and in 247 of 432 patients (57%) who received placebo (HRrelapse or death, 0.47; 95% CI, 0.39–0.58; P < .001). Median RFS had not been reached in the combination arm (95% CI, 44.5–not reached) and was 16.6 months (95% CI, 12.7–22.1) in the placebo group.[26][Level of evidence B1]
- In the combination therapy arm, 26% of the patients had an adverse event leading to the discontinuation of therapy, 38% required dose reduction, and 66% required dose interruption. In the placebo arm, 3% of the patients had an adverse event leading to discontinuation of therapy, 3% required dose reduction, and 15% required dose interruption. Serious adverse events occurred in 36% of the patients who received the combination therapy and 10% of the patients in the placebo group. One death, which resulted from pneumonia, was reported in the combination therapy arm.
- A final analysis performed after a minimum of 8.33 years of follow-up showed superior RFS for patients who received the dabrafenib plus trametinib combination compared with patients who received placebo. Of patients who received the combination, 52% (95% CI, 47.4%–56.3%) were alive without relapse compared with 36% (95% CI, 30.9%–40.2%) of patients who received the placebo (HR, 0.52; 95% CI, 0.43–0.63). DMFS was also improved in patients who received the combination (60% vs. 46%; HR, 0.56; 95% CI, 0.44–0.71). An OS analysis showed improvement that was not statistically significant (HR, 0.80; 95% CI, 0.62–1.01).[31,32]
Current Clinical Trials
Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria. General information about clinical trials is also available.
References
- Veronesi U, Cascinelli N: Narrow excision (1-cm margin). A safe procedure for thin cutaneous melanoma. Arch Surg 126 (4): 438-41, 1991. [PUBMED Abstract]
- Veronesi U, Cascinelli N, Adamus J, et al.: Thin stage I primary cutaneous malignant melanoma. Comparison of excision with margins of 1 or 3 cm. N Engl J Med 318 (18): 1159-62, 1988. [PUBMED Abstract]
- Wagner JD, Gordon MS, Chuang TY, et al.: Current therapy of cutaneous melanoma. Plast Reconstr Surg 105 (5): 1774-99; quiz 1800-1, 2000. [PUBMED Abstract]
- Cohn-Cedermark G, Rutqvist LE, Andersson R, et al.: Long term results of a randomized study by the Swedish Melanoma Study Group on 2-cm versus 5-cm resection margins for patients with cutaneous melanoma with a tumor thickness of 0.8-2.0 mm. Cancer 89 (7): 1495-501, 2000. [PUBMED Abstract]
- Balch CM, Soong SJ, Smith T, et al.: Long-term results of a prospective surgical trial comparing 2 cm vs. 4 cm excision margins for 740 patients with 1-4 mm melanomas. Ann Surg Oncol 8 (2): 101-8, 2001. [PUBMED Abstract]
- Heaton KM, Sussman JJ, Gershenwald JE, et al.: Surgical margins and prognostic factors in patients with thick (>4mm) primary melanoma. Ann Surg Oncol 5 (4): 322-8, 1998. [PUBMED Abstract]
- Balch CM, Urist MM, Karakousis CP, et al.: Efficacy of 2-cm surgical margins for intermediate-thickness melanomas (1 to 4 mm). Results of a multi-institutional randomized surgical trial. Ann Surg 218 (3): 262-7; discussion 267-9, 1993. [PUBMED Abstract]
- Shen P, Wanek LA, Morton DL: Is adjuvant radiotherapy necessary after positive lymph node dissection in head and neck melanomas? Ann Surg Oncol 7 (8): 554-9; discussion 560-1, 2000. [PUBMED Abstract]
- Hochwald SN, Coit DG: Role of elective lymph node dissection in melanoma. Semin Surg Oncol 14 (4): 276-82, 1998. [PUBMED Abstract]
- Cascinelli N, Morabito A, Santinami M, et al.: Immediate or delayed dissection of regional nodes in patients with melanoma of the trunk: a randomised trial. WHO Melanoma Programme. Lancet 351 (9105): 793-6, 1998. [PUBMED Abstract]
- Koops HS, Vaglini M, Suciu S, et al.: Prophylactic isolated limb perfusion for localized, high-risk limb melanoma: results of a multicenter randomized phase III trial. European Organization for Research and Treatment of Cancer Malignant Melanoma Cooperative Group Protocol 18832, the World Health Organization Melanoma Program Trial 15, and the North American Perfusion Group Southwest Oncology Group-8593. J Clin Oncol 16 (9): 2906-12, 1998. [PUBMED Abstract]
- Wong SL, Balch CM, Hurley P, et al.: Sentinel lymph node biopsy for melanoma: American Society of Clinical Oncology and Society of Surgical Oncology joint clinical practice guideline. J Clin Oncol 30 (23): 2912-8, 2012. [PUBMED Abstract]
- Kirkwood JM, Strawderman MH, Ernstoff MS, et al.: Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J Clin Oncol 14 (1): 7-17, 1996. [PUBMED Abstract]
- Kirkwood JM, Ibrahim JG, Sondak VK, et al.: High- and low-dose interferon alfa-2b in high-risk melanoma: first analysis of intergroup trial E1690/S9111/C9190. J Clin Oncol 18 (12): 2444-58, 2000. [PUBMED Abstract]
- Eggermont AM, Suciu S, Santinami M, et al.: Adjuvant therapy with pegylated interferon alfa-2b versus observation alone in resected stage III melanoma: final results of EORTC 18991, a randomised phase III trial. Lancet 372 (9633): 117-26, 2008. [PUBMED Abstract]
- Hancock BW, Wheatley K, Harris S, et al.: Adjuvant interferon in high-risk melanoma: the AIM HIGH Study--United Kingdom Coordinating Committee on Cancer Research randomized study of adjuvant low-dose extended-duration interferon Alfa-2a in high-risk resected malignant melanoma. J Clin Oncol 22 (1): 53-61, 2004. [PUBMED Abstract]
- Chapman PB, Hauschild A, Robert C, et al.: Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364 (26): 2507-16, 2011. [PUBMED Abstract]
- Faries MB, Thompson JF, Cochran AJ, et al.: Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N Engl J Med 376 (23): 2211-2222, 2017. [PUBMED Abstract]
- Leiter U, Stadler R, Mauch C, et al.: Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol 17 (6): 757-767, 2016. [PUBMED Abstract]
- Patel SP, Othus M, Chen Y, et al.: Neoadjuvant-Adjuvant or Adjuvant-Only Pembrolizumab in Advanced Melanoma. N Engl J Med 388 (9): 813-823, 2023. [PUBMED Abstract]
- Blank CU, Lucas MW, Scolyer RA, et al.: Neoadjuvant Nivolumab and Ipilimumab in Resectable Stage III Melanoma. N Engl J Med 391 (18): 1696-1708, 2024. [PUBMED Abstract]
- Eggermont AM, Chiarion-Sileni V, Grob JJ, et al.: Prolonged Survival in Stage III Melanoma with Ipilimumab Adjuvant Therapy. N Engl J Med 375 (19): 1845-1855, 2016. [PUBMED Abstract]
- Tarhini AA, Lee SJ, Hodi FS, et al.: Phase III Study of Adjuvant Ipilimumab (3 or 10 mg/kg) Versus High-Dose Interferon Alfa-2b for Resected High-Risk Melanoma: North American Intergroup E1609. J Clin Oncol 38 (6): 567-575, 2020. [PUBMED Abstract]
- Weber J, Mandala M, Del Vecchio M, et al.: Adjuvant Nivolumab versus Ipilimumab in Resected Stage III or IV Melanoma. N Engl J Med 377 (19): 1824-1835, 2017. [PUBMED Abstract]
- Eggermont AMM, Blank CU, Mandala M, et al.: Adjuvant Pembrolizumab versus Placebo in Resected Stage III Melanoma. N Engl J Med 378 (19): 1789-1801, 2018. [PUBMED Abstract]
- Long GV, Hauschild A, Santinami M, et al.: Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med 377 (19): 1813-1823, 2017. [PUBMED Abstract]
- Maio M, Lewis K, Demidov L, et al.: Adjuvant vemurafenib in resected, BRAFV600 mutation-positive melanoma (BRIM8): a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol 19 (4): 510-520, 2018. [PUBMED Abstract]
- Ascierto PA, Del Vecchio M, Mandalá M, et al.: Adjuvant nivolumab versus ipilimumab in resected stage IIIB-C and stage IV melanoma (CheckMate 238): 4-year results from a multicentre, double-blind, randomised, controlled, phase 3 trial. Lancet Oncol 21 (11): 1465-1477, 2020. [PUBMED Abstract]
- Eggermont AMM, Blank CU, Mandalà M, et al.: Adjuvant pembrolizumab versus placebo in resected stage III melanoma (EORTC 1325-MG/KEYNOTE-054): distant metastasis-free survival results from a double-blind, randomised, controlled, phase 3 trial. Lancet Oncol 22 (5): 643-654, 2021. [PUBMED Abstract]
- Eggermont AM, Chiarion-Sileni V, Grob JJ, et al.: Adjuvant ipilimumab versus placebo after complete resection of high-risk stage III melanoma (EORTC 18071): a randomised, double-blind, phase 3 trial. Lancet Oncol 16 (5): 522-30, 2015. [PUBMED Abstract]
- Dummer R, Hauschild A, Santinami M, et al.: Five-Year Analysis of Adjuvant Dabrafenib plus Trametinib in Stage III Melanoma. N Engl J Med 383 (12): 1139-1148, 2020. [PUBMED Abstract]
- Long GV, Hauschild A, Santinami M, et al.: Final Results for Adjuvant Dabrafenib plus Trametinib in Stage III Melanoma. N Engl J Med 391 (18): 1709-1720, 2024. [PUBMED Abstract]
Treatment of Unresectable Stage III, Stage IV, and Recurrent Melanoma
Treatment Options for Unresectable Stage III, Stage IV, and Recurrent Melanoma
Treatment options for unresectable stage III, stage IV, and recurrent melanoma include the following:
- Immunotherapy.
- Signal transduction inhibitors.
- BRAF inhibitors (for patients who test positive for the BRAF V600 pathogenic variant).
- Mitogen-activated protein kinase (MEK) inhibitors.
- KIT inhibitors.
- Combination therapy with signal transduction inhibitors.
- Combination signal transduction inhibitor therapy plus PD-L1 inhibitor.
- Tumor-infiltrating lymphocyte (TIL) therapy.
- Intralesional therapy.
- Adjunctive local/regional therapy including surgical resection.
- Isolated limb infusion (ILI).
- Palliative therapy.
- Targeted therapy with single agents or combination therapy (under clinical evaluation).
- Combinations of immunotherapy and targeted therapy (under clinical evaluation).
- Intralesional injections (e.g., oncolytic viruses) (under clinical evaluation).
- Complete surgical resection of all known disease versus best medical therapy (under clinical evaluation).
- Isolated limb perfusion for unresectable extremity melanoma (under clinical evaluation).
- Systemic therapy for unresectable disease (under clinical evaluation).
Two approaches—checkpoint inhibition and targeting the mitogen-activated protein kinase (MAPK) pathway—improved progression-free survival (PFS) and overall survival (OS) in randomized trials. Anti–PD-1 monotherapy (pembrolizumab or nivolumab) improved efficacy outcomes with better safety profiles when compared with treatment using single-agent anti–CTLA-4 (ipilimumab) or investigator choice of chemotherapy. The combination of anti–PD-1 and anti–CTLA-4 immunotherapies (nivolumab and ipilimumab) also prolongs PFS and OS compared with ipilimumab, but the combination is associated with significant toxicity. The efficacy seen with immunotherapy is independent of BRAF variant status.
Combinations of BRAF and MEK inhibitors have consistently shown superior efficacy compared with BRAF monotherapy. Improved PFS was seen when a PD-L1 inhibitor (atezolizumab) was added to the combination of a BRAF plus MEK inhibitor (vemurafenib plus cobimetinib); however, data on OS are immature. Further questions remain regarding triplet therapy, including how it compares with monotherapy checkpoint inhibition and if the concurrent administration is superior to sequential therapy (NCT02224781).
Because of the rapid development of new agents, combinations, and remaining questions, patients and their physicians are encouraged to consider a clinical trial for initial treatment and at the time of progression. Ongoing clinical trials address the following issues:
- The value of sequencing therapies, such as immunotherapy and targeted therapy.
- Optimal doses of combination immunotherapy to decrease toxicity and preserve efficacy.
- How to select the patients who will benefit from combination immunotherapy versus monotherapy.
- The role of PD-L1 expression as a biomarker for efficacy.
- The role of maintenance therapy.
Immunotherapy
Checkpoint inhibitors
Anti–PD-1 and PD-L1 therapy
The PD-1 pathway is a key immunoinhibitory mediator of T-cell exhaustion. Blockade of this pathway can lead to T-cell activation, expansion, and enhanced effector functions. PD-1 has two ligands, PD-L1 and PD-L2. The U.S. Food and Drug Administration (FDA) has approved two anti–PD-1 antibodies, pembrolizumab and nivolumab, based on improved OS in randomized trials.
Pembrolizumab
Evidence (pembrolizumab):
- Previously treated patients. One study included 173 patients with unresectable or metastatic melanoma with disease progression within 24 weeks of the last dose of ipilimumab. If the patient had a BRAF V600 pathogenic variant, previous treatment with a BRAF inhibitor was also required. Patients were randomly assigned to one of two doses of pembrolizumab—2 mg/kg or 10 mg/kg—every 3 weeks. The trial excluded patients with an autoimmune disease, a condition requiring immunosuppression, or a history of severe immune-related adverse events (irAEs) from treatment with ipilimumab.
- The median age was 61 years; 60% were male; 67% had an Eastern Cooperative Oncology Group (ECOG) performance status (PS) score of 0, and 33% had an ECOG PS of 1. Eighteen percent of patients had BRAF V600 pathogenic variants, 39% had an elevated lactate dehydrogenase (LDH) level, 64% had M1c disease, 9% had brain metastases, and 72% had undergone two or more therapies for advanced disease. The primary outcome measure was overall response rate according to Response Evaluation Criteria In Solid Tumors (RECIST, version 1.1) criteria as assessed by blinded independent central review.[1][Level of evidence B3]
- The overall response rate was 26% (95% confidence interval [CI], -14 to 13; P = .96) for patients who received the 2 mg/kg dose, consisting of one complete response and 20 partial responses in 81 patients. Median follow-up was 8 months, and all patients had a minimum of 6 months of follow-up. Among the 21 patients with an objective response, 18 had ongoing responses, ranging from 1.4+ months to 8.5+ months.
- Patients who received the 10 mg/kg dose had a similar response rate (26%), consisting of 20 responses in 76 patients. Responses were seen in patients with and without BRAF V600 pathogenic variants.
- The approved dose was 2 mg/kg given as an intravenous (IV) infusion for 30 minutes every 3 weeks.
Pembrolizumab was discontinued because of adverse events in 7% of the patients treated with 2 mg/kg, with 3% considered drug-related adverse events by the investigators. The most common adverse events were the following (2 mg/kg vs. 10 mg/kg arms):
- Fatigue (33% vs. 37%).
- Pruritus (23% vs. 19%).
- Rash (18% vs. 18%).
Other common adverse events included cough, nausea, decreased appetite, constipation, arthralgia, and diarrhea. The most frequent and serious adverse events that occurred in more than 2% of the 411 patients treated with pembrolizumab included renal failure, dyspnea, pneumonia, and cellulitis. Additional clinically significant irAEs included pneumonitis, colitis, hypophysitis, hyperthyroidism, hypothyroidism, nephritis, and hepatitis.
The FDA label provides recommendations for suspected irAEs, including withholding the drug and administering corticosteroids.
- Previously untreated and treated patients. A multicenter international trial (KEYNOTE 006 [NCT01866319]) randomly assigned 834 patients with metastatic melanoma in a 1:1:1 ratio to receive pembrolizumab (10 mg/kg IV every 2 weeks or every 3 weeks) or four cycles of ipilimumab (3 mg/kg every 3 weeks).[2] Patients were stratified by ECOG PS (0 vs. 1), line of therapy (first-line vs. second-line), and PD-L1 expression (positive vs. negative). The primary end points were PFS and OS.[2][Level of evidence A1]
Approximately 66% of patients had received no previous systemic therapy for advanced melanoma. BRAF V600 pathogenic variants were present in 36% of patients and of these, approximately 50% had received previous BRAF inhibitor treatments. The study did not enroll patients with BRAF V600 pathogenic variants with high LDH levels and symptomatic or rapidly progressive disease who had not received anti-BRAF therapy, which could provide rapid clinical benefit. Approximately 80% of patients had PD-L1–positive tissue samples.
- The final protocol-specified analysis of OS was conducted at a median follow-up of 23 months. Median OS was not reached in either pembrolizumab group; however, OS was 16.0 months for the ipilimumab group (hazard ratio [HR], 0.68; 95% CI, 0.53−0.87 for pembrolizumab every 2 weeks vs. ipilimumab; P = .0009 and 0.68; 95% CI, 0.53−0.86 for pembrolizumab every 3 weeks vs. ipilimumab; P = .0008). The 24-month survival rate was 55% in the groups who received pembrolizumab every 2 weeks and every 3 weeks compared with 43% in the ipilimumab group.[3]
- Benefit was seen across all subgroups except for patients with PD-L1–negative tumors. However, since this subset was small (18% of patients) and the CI was wide, no definitive conclusions could be drawn from this study.
Nivolumab
Evidence (nivolumab):
- Previously treated patients. Accelerated approval was based on a planned noncomparative interim analysis of the first 120 patients who received nivolumab with at least 6 months of follow-up from a multicenter, open-label trial (CheckMate 037 [NCT01721746]). In this trial, patients were randomly assigned (2:1) to receive either nivolumab (3 mg/kg every 2 weeks) or the investigator’s choice of chemotherapy (either dacarbazine 1,000 mg/m2 IV every 3 weeks or the combination of carboplatin [area under the curve 6] every 3 weeks plus paclitaxel 175 mg/m2 every 3 weeks).[FDA label][Level of evidence C3] Patients were required to have unresectable or metastatic melanoma that had progressed after treatment with ipilimumab. If the patient had a BRAF V600 pathogenic variant, previous treatment with a BRAF inhibitor was also required. The trial excluded patients with an autoimmune disease, a condition requiring immunosuppression, or a history of severe irAEs from treatment with ipilimumab.
- The median age of patients was 58 years; 65% of patients were male; and ECOG PS was 0 in 58% of patients. BRAF V600 pathogenic variants were present in 22% of patients; 76% had M1c disease; 56% had elevated LDH levels; 18% had a history of brain metastases; and, 68% had previously received two or more systemic therapies for metastatic disease.
- Objective response rate and OS were coprimary end points. The objective response rate was 32% (95% CI, 23%–41%), with four complete responses and 34 partial responses as assessed by RECIST 1.1 criteria and an independent central review. Among the 38 patients with responses, 33 (87%) had ongoing responses, with durations from 2.6+ to 10.0+ months.
- Responses were seen in patients with and without BRAF V600 pathogenic variants.
- Safety analysis is based on 268 patients. Nivolumab was discontinued because of adverse events in 9% of patients. Serious adverse events occurred in 41% of patients, and grade 3 and grade 4 adverse events occurred in 42% of patients. The most common adverse events were rash, cough, upper respiratory tract infection, and peripheral edema. Other important adverse events included ventricular arrhythmia, iridocyclitis, increased amylase and lipase, dizziness, and neuropathy.
The FDA label provides recommendations for suspected irAEs, including withholding the drug and administering corticosteroids.
- Previously untreated patients. A double-blind multicenter trial (CheckMate 066 [NCT01721772]) included 418 patients with unresectable stage III or stage IV melanoma without BRAF pathogenic variants. Patients were randomly assigned (1:1) to receive either nivolumab (3 mg/kg every 2 weeks) and a dacarbazine-matched placebo (every 3 weeks) or dacarbazine (1,000 mg/m2 every 3 weeks with a nivolumab-matched placebo every 2 weeks). The primary end point was OS.[4][Level of evidence A1] The trial was conducted in 80 centers in Europe, Israel, Australia, Canada, and South America, which are countries where dacarbazine had been a standard first-line treatment in patients without BRAF pathogenic variants.
- The Data and Monitoring Safety Board (DMSB) noted a potential difference in OS during safety review. In 2014, an abbreviated report from an unplanned interim–database lock was reviewed showing a significant difference in OS, in favor of nivolumab. The DMSB recommended that the study be unblinded and allow patients on dacarbazine to receive nivolumab. The intended sample size was approximately 410 patients; a total of 418 patients had been entered.
- Results from the double-blind portion of the study before the crossover amendment showed that median OS was not reached in the nivolumab group and was 10.8 months (95% CI, 9.3–12.1) in the dacarbazine group. The 1-year OS rate was 72.9% (95% CI, 65.%–78.9%) in the nivolumab group and 42.1% (95% CI, 33.0%–50.9%) in the dacarbazine group. The HRdeath was 0.42 (99.79% CI, 0.25–0.73; P < .001).
- The most common adverse events in the nivolumab group were fatigue (19.9%), pruritus (17%), nausea (16.5%), and diarrhea (16%). Adverse events led to treatment discontinuation in 6.8% of patients in the nivolumab group and 11.7% of patients in the dacarbazine group. Adverse events with potential immunological etiology that occurred included gastrointestinal, hepatic, pulmonary, renal, endocrine, and skin. However, most resolved with a delay in study treatment, glucocorticoid administration, or both per management guidelines for nivolumab. No deaths were attributed to drug-related adverse events in either group.
-
Change in dosing regimen for nivolumab in metastatic melanoma.
- In a population pharmacokinetic response analysis and a dose/exposure-response analysis, the flat dose of 240 mg of nivolumab every 2 weeks was considered pharmacokinetically equivalent to the dosing regimen of 3 mg/kg. Clinical safety and efficacy at the two doses appeared similar across body weight and tumor types in melanoma, non-small cell lung cancer, and renal cell carcinoma.[5]
- The dosing regimen approved by the FDA for monotherapy has changed from 3 mg/kg to 240 mg IV every 2 weeks until disease progression or intolerable toxicity. The dosing regimen of 1 mg/kg of IV nivolumab when combined with ipilimumab will remain unchanged until after therapy with ipilimumab is complete, when the regimen will change to a 240 mg dose every 2 weeks until disease progression or intolerable toxicity.
Anti–cytotoxic T-lymphocyte antigen-4 (CTLA-4) therapy
Ipilimumab
Ipilimumab is a human monoclonal antibody that binds to CTLA-4, thereby blocking its ability to downregulate T-cell activation, proliferation, and effector function.
Approved by the FDA in 2011, ipilimumab has demonstrated clinical benefit by prolonging OS in randomized trials. Two prospective, randomized, international trials, one each in previously untreated and treated patients, supported the use of ipilimumab.[6,7]
Evidence (ipilimumab):
- Previously treated patients. A total of 676 patients with previously treated, unresectable stage III or stage IV disease, and who were HLA-A*0201-positive, were enrolled in a three-arm, multinational, randomized (3:1:1), double-blind, double-placebo trial. A total of 403 patients were randomly assigned to receive ipilimumab (3 mg/kg every 3 weeks for 4 doses) with glycoprotein 100 (gp100) peptide vaccine. One hundred thirty-seven patients received ipilimumab (3 mg/kg every 3 weeks for 4 doses), and 136 patients received the gp100 vaccine. Patients were stratified by baseline metastases and previous receipt or nonreceipt of IL-2 therapy. Eighty-two of the patients had metastases to the brain at baseline.[7][Level of evidence A1]
- The median OS was 10 months in patients who received ipilimumab alone and 10.1 months in those who received ipilimumab with the gp100 vaccine, compared with 6.4 months for patients who received the vaccine alone (HR of ipilimumab alone vs. gp100 alone, 0.66; P < .003; HR of ipilimumab plus vaccine vs. gp100 alone, 0.68; P < .001).
- An analysis at 1 year showed that 44% of patients who were treated with ipilimumab and 45% of those treated with ipilimumab and the vaccine were alive, compared with 25% of the patients who received the vaccine only.
- Grade 3 or grade 4 irAEs occurred in 10% to 15% of patients treated with ipilimumab. These irAEs most often included diarrhea or colitis, and endocrine-related events (e.g., inflammation of the pituitary). These events required cessation of therapy and institution of anti-inflammatory agents such as corticosteroids or, in four cases, infliximab (an antitumor necrosis factor-alpha antibody).
- There were 14 drug-related deaths (2.1%), and seven deaths were associated with irAEs.
- Previously untreated patients. A multicenter, international trial included 502 patients who were untreated for metastatic disease (adjuvant treatment was allowed). Patients were randomly assigned (1:1) to receive either ipilimumab (10 mg/kg) plus dacarbazine (850 mg/m2) or placebo plus dacarbazine (850 mg/m2) at weeks 1, 4, 7, and 10 followed by dacarbazine alone every 3 weeks through week 22. Patients with stable disease or an objective response and no dose-limiting toxic effects received ipilimumab or placebo every 12 weeks thereafter as maintenance therapy. The primary end point was survival. Patients were stratified according to ECOG PS and metastatic stage. Approximately 70% of the patients had an ECOG PS of 0, and the remainder of the patients had an ECOG PS of 1. Approximately 55% of patients had stage M1c disease.[6][Level of evidence A1]
- The median OS was 11.2 months (95% CI, 9.4–13.6) in the ipilimumab-dacarbazine group versus 9.1 months (95% CI, 7.8–10.5) in the placebo-dacarbazine group. In the ipilimumab-dacarbazine group, estimated survival rates were 47.3% at 1 year, 28.5% at 2 years, and 20.8% at 3 years (HRdeath, 0.72; P < .001). In the placebo-dacarbazine group, the survival rates were 36.3% at 1 year, 17.9% at 2 years, and 12.2% at 3 years.
- The most common study-drug–related adverse events were those classified as immune related. Grade 3 or grade 4 irAEs were seen in 38.1% of patients treated with ipilimumab plus dacarbazine versus 4.4% of patients treated with placebo plus dacarbazine. The most common events were hepatitis and enterocolitis.
- No drug-related deaths occurred.
Clinicians and patients should be aware that immune-mediated adverse reactions may be severe or fatal. Early identification and treatment are necessary, including potential administration of systemic glucocorticoids or other immunosuppressants according to the immune-mediated adverse reaction management guide provided by the manufacturer.[8]
High-dose IL-2
The FDA approved IL-2 in 1998 because of durable complete responses in eight phase I and II studies. Phase III trials comparing high-dose IL-2 to other re-treatments, providing an assessment of relative impact on OS, have not been conducted.
Evidence (high-dose IL-2):
- Based on a pooled analysis of 270 patients from eight single- and multi-institutional trials in 22 institutions conducted between 1985 and 1993, results included:
Strategies to improve this therapy are an active area of investigation.
Dual checkpoint inhibition
T cells coexpress several receptors that inhibit T-cell function. Preclinical data and early clinical data suggest that co-blockade of two inhibitory receptors may be more effective than blockade of either alone.[11] This led to a phase III trial (NCT01844505) comparing each single agent with the combination of ipilimumab and nivolumab, and another phase III trial comparing nivolumab to the combination of nivolumab and relatlimab (NCT03470922). The FDA has approved both the ipilimumab-nivolumab and the nivolumab-relatlimab combinations.
CTLA-4 inhibitor plus PD-1 inhibitor
Evidence (ipilimumab plus nivolumab):
- Previously untreated patients. In an international double-blind trial (CheckMate 067), 945 previously untreated patients with unresectable stage III or IV melanoma were randomly assigned in a 1:1:1 ratio to receive one of the following regimens:
- Arm 1: nivolumab (3 mg/kg every 2 weeks) plus placebo;
- Arm 2: nivolumab (1 mg/kg every 3 weeks) plus ipilimumab (3 mg/kg every 3 weeks for 4 doses) followed by nivolumab (3 mg/kg every 2 weeks for cycle 3 and beyond); or
- Arm 3: ipilimumab (3 mg/kg every 3 weeks for 4 doses) plus placebo.
PFS and OS were coprimary end points. The study was powered to compare the combination of nivolumab plus ipilimumab with ipilimumab monotherapy, and nivolumab monotherapy with ipilimumab monotherapy. The study was not powered to compare combination ipilimumab plus nivolumab with nivolumab.
Patients were stratified according to tumor PD-L1 status assessed in a central laboratory by immunohistochemical testing (positive vs. negative or indeterminate), BRAF pathogenic variant status (V600 variant−positive vs. wild-type), and American Joint Committee on Cancer stage. Characteristics at baseline were as follows: 74% of patients had an ECOG PS of 0; 36% had elevated LDH levels; 31.5% had BRAF pathogenic variants; and 58% had M1c disease. A minority of patients (23.6%) had PD-L1–positive tumors.[12][Level of evidence A1]
- The prospectively defined coprimary analysis of PFS occurred after all patients had at least 9 months of follow-up. Treatment with nivolumab alone or in combination with ipilimumab resulted in significantly longer PFS than with ipilimumab alone. Results were consistent across the prespecified stratification factors. Median PFS was 6.9 months (95% CI, 4.3–9.5) with nivolumab, 11.5 months (95% CI, 8.9–16.7) with nivolumab plus ipilimumab, and 2.9 months (95% CI, 2.8–3.4) with ipilimumab.
- The prospectively specified coprimary analysis of OS was to occur at 28 months. With 467 deaths, the OS rate at this time point was 59% in the nivolumab group, 64% in the combination group, and 45% in the ipilimumab group (HRdeath for the combination vs. ipilimumab, 0.55 [98% CI, 0.42–0.72; P< .001]; HRdeath with nivolumab vs. ipilimumab 0.63 [98% CI, 0.48–0.81; P< .001]).[13]
- In a descriptive analysis with a minimum follow-up of 36 months, the following data were found:
- OS rates were 52% in the nivolumab group, 58% in the combination group, and 34% in the ipilimumab group.
- The median OS was not reached in the combination arm (95% CI, 38.2 months–not reached). Median OS in the single-agent nivolumab and ipilimumab groups were 37.6 months (95% CI, 29.1–not reached) and 19.9 months (95% CI, 16.9–24.6), respectively.
- For the combination versus ipilimumab, the HRdeath was 0.55 (99.5% CI, 0.45–0.69; P < .001); for nivolumab versus ipilimumab, the HR was 0.65 (99.5% CI, 0.53–0.80; P < .001).
- Adverse events were highest in the combination arm and need to be monitored carefully. Grades 3 to 4 treatment-related adverse events occurred in 16.3% of patients in the nivolumab group, 27.3% of patients in the ipilimumab group, and 55% of patients in the combination group. The most frequent reason for treatment discontinuation was disease progression in the two monotherapy arms—49% with nivolumab and 65% with ipilimumab. The most frequent reason for discontinuation in the combination group was toxicity (38%).
- Four therapy-related deaths were reported, which were attributed to neutropenia (nivolumab group), colon perforation (ipilimumab group), liver necrosis, and autoimmune myocarditis (combination ipilimumab and nivolumab).
- Analyses of PD-L1 expression level associated with OS at 3 years showed that expression levels alone are not a reliable predictor of OS.
- In a final analysis with a minimum follow-up of 10 years, the following data were reported:
- The 10-year OS rate was 34% in the combination group, 26% in the nivolumab group, and 16% in the ipilimumab group.
- The median OS was 71.9 months (95% CI, 51.6–not reached) in the combination group, 36.9 months (95% CI, 30.7–46.6) in the nivolumab group, and 19.9 months (95% CI, 16.8–24.6) in the ipilimumab group.
- HRdeath, compared with the ipilimumab group, was 0.53 (95% CI, 0.44–0.65) for the combination group and 0.70 (95% CI, 0.59–0.84) for the nivolumab group.[14]
- Melanoma metastatic to the brain. Patients with at least one measurable, nonirradiated brain metastasis were eligible for treatment with systemic dual immunotherapy in an open-label, multicenter phase II trial (CheckMate 204 [NCT02320058]).[15][Level of evidence C3] Eligibility required no need for immediate intervention, an absence of neurological signs or symptoms, and no glucocorticoids within 14 days of study treatment. Patients may have received previous stereotactic radiosurgery or excision of up to three brain metastases. Positive PD-L1 expression was not required.
Treatment consisted of nivolumab (1 mg/kg) plus ipilimumab (3 mg/kg) every 3 weeks for up to 4 doses, followed by nivolumab (3 mg/kg) every 2 weeks until progression or unacceptable toxicity.
The primary end point was rate of intracranial clinical benefit assessed by the investigator per RECIST criteria and defined as the percentage of patients with a complete response, partial response, or stable disease for at least 6 months. A total of 28 sites in the United States enrolled 101 patients, 94 of whom had a minimum follow-up of 6 months; the data on that population are reported below.
- Clinical benefit (in the brain) was seen in 57% of patients (95% CI, 47%–68%); 24 patients (26%) had a complete response, 28 patients (30%) had a partial response, and 2 patients (2%) had stable disease that lasted for 6 months or longer. Similar rates of objective response (50%) were seen in patients with extracranial lesions, although fewer patients had a complete response (7%).
- A subgroup analysis indicated responses in both PD-L1–positive and PD-L1–negative patients (baseline status not known in 20/94 patients).
- The median follow-up of the 94 patients was 14 months. Median time to intracranial response was 2.3 months (range, 1.1−10.8), and time to extracranial response was 2.1 months (range, 1.1−15.0).
- The most common treatment-related adverse event of any grade in the nervous system was headache (21 patients [22%]), with 3 patients (3%) having headaches of grade 3 or 4. Other treatment-related neurological adverse events of grade 3 or 4 were brain edema (2 patients [2%]), intracranial hemorrhage (1 patient [1%]), peripheral motor neuropathy (1 patient [1%]), and syncope (1 patient [1%]). Each of these adverse events led to treatment discontinuation, and the one reported case of peripheral motor neuropathy was irreversible. The investigator determined one death to be related to the study treatment (grade 5 immune-related myocarditis).
- Progression was documented in 33 patients (35%); 17 patients (18%) had intracranial progression only, 4 patients (4%) had extracranial progression only, and 12 patients (13%) had progression in both intracranial and extracranial sites.
LAG-3 inhibitor plus PD-1 inhibitor
Evidence (relatlimab plus nivolumab):
- A multinational, phase II/III, double-blind trial (RELATIVITY-047 [NCT03470922]) included 714 patients with previously untreated, histologically confirmed, unresectable stage III or IV melanoma. Patients were randomly assigned in a 1:1 ratio to receive either 160 mg of relatlimab and 480 mg of nivolumab in a fixed-dose combination (n = 355) or 480 mg of nivolumab (n = 359). The characteristics of the patients at baseline were well balanced between the treatment groups. Both therapies were given in a single 60-minute IV infusion every 4 weeks. The primary end point was PFS assessed according to RECIST, version 1.1. Secondary end points included OS and objective response. Patients who had received previous adjuvant or neoadjuvant therapies containing a PD-1, CTLA-4, BRAF, or MEK inhibitor (or a combination of BRAF and MEK inhibitors) were eligible if the therapy was completed at least 6 months before the date of recurrence. Key exclusion criteria were uveal melanoma and active, untreated brain or leptomeningeal metastases.[16]
- The median PFS was 10.1 months (95% CI, 6.4–15.7) in the relatlimab-nivolumab group and 4.6 months (95% CI, 3.4–5.6) in the nivolumab-alone group (HRprogression or death, 0.75; 95% CI, 0.62–0.92; P = .006). The 12-month PFS rate was 47.7% (95% CI, 41.8%–53.2%) in the relatlimab-nivolumab group and 36.0% (95% CI, 30.5%–41.6%) in the nivolumab-alone group.[16][Level of evidence B1]
- Grade 3 or 4 treatment-related adverse events occurred in 18.9% of the patients in the relatlimab-nivolumab group and 9.7% of patients in the nivolumab-alone group. Treatment-related adverse events led to treatment discontinuation in 14.6% of patients in the relatlimab-nivolumab group and 6.7% of patients in the nivolumab-alone group. The most common categories of immune-mediated adverse events that occurred in the relatlimab-nivolumab group were hypothyroidism or thyroiditis (18.0%), rash (9.3%), and diarrhea or colitis (6.8%). Myocarditis occurred in 1.7% of the patients in the relatlimab-nivolumab group and 0.6% of patients in the nivolumab-alone group.
- The relatlimab-nivolumab combination was beneficial compared with nivolumab alone, regardless of LAG-3 expression or BRAF pathogenic variant status. OS and response rate data have not been reported.
Signal transduction inhibitors
Studies indicate that both BRAF and MEK inhibitors, as single agents and in combination, can significantly impact the natural history of melanoma, although they do not appear to provide a cure.
BRAF inhibitors
Treatment with BRAF inhibitors is discouraged for patients with wild-type BRAF melanoma because data from preclinical models have demonstrated that BRAF inhibitors can enhance rather than downregulate the MAPK pathway in tumor cells with wild-type BRAF and upstream RAS pathogenic variants.[17-20]
Vemurafenib
Vemurafenib is an orally available, small-molecule, selective BRAF kinase inhibitor that was approved by the FDA in 2011 for patients with unresectable or metastatic melanoma and BRAF V600E pathogenic variants.
Evidence (vemurafenib):
- Previously untreated patients. The approval of vemurafenib was supported by an international multicenter trial (BRIM-3 [NCT01006980]) that screened 2,107 patients with previously untreated stage IIIC or IV melanoma for BRAF V600 pathogenic variants and identified 675 patients via the cobas 4800 BRAF V600 Mutation Test.[21] Patients were randomly assigned to receive either vemurafenib (960 mg by mouth twice a day) or dacarbazine (1,000 mg/m2 IV every 3 weeks). Coprimary end points were rates of OS and PFS. At the planned interim analysis, the DMSB determined that both the OS and PFS end points had met the prespecified criteria for statistical significance in favor of vemurafenib and recommended that patients in the dacarbazine group be allowed to cross over to receive vemurafenib.[21][Levels of evidence A1 and B1]
- A total of 675 patients were evaluated for OS. Although the median survival had not yet been reached for vemurafenib and the data were immature for reliable Kaplan-Meier estimates of survival curves, the OS in the vemurafenib arm was clearly superior to that in the dacarbazine arm.
- The HRdeath in the vemurafenib group was 0.37 (95% CI, 0.26–0.55; P < .001). The survival benefit in the vemurafenib group was observed in each prespecified subgroup, for example, age, sex, ECOG PS, tumor stage, LDH level, and geographical region.
- The HR for tumor progression was 0.26 (95% CI, 0.20–0.33; P < .001) in the vemurafenib arm. The estimated median PFS was 5.3 months in the vemurafenib arm versus 1.6 months in the dacarbazine arm.
- Twenty patients had non-BRAF V600E pathogenic variants: 19 with BRAF V600K and 1 with BRAF V600D. Four patients with BRAF V600K pathogenic variants had a response to vemurafenib.
- Adverse events required dose modification or interruption in 38% of patients who received vemurafenib and 16% of those who received dacarbazine. The most common adverse events with vemurafenib were cutaneous events, arthralgia, and fatigue. Cutaneous squamous cell carcinoma (SCC), keratoacanthoma, or both developed in 18% of patients and were treated by simple excision. The most common adverse events with dacarbazine were fatigue, nausea, vomiting, and neutropenia. For more information, visit Fatigue and Nausea and Vomiting Related to Cancer Treatment.
- Previously treated patients. A multicenter phase II trial included 132 patients with BRAF V600E or BRAF V600K pathogenic variants. Patients received vemurafenib (960 mg by mouth twice a day). Of the enrolled patients, 61% had stage M1c disease, and 49% had an elevated LDH level. All patients had received one or more previous therapies for advanced disease. Median follow-up was 12.9 months.[22][Level of evidence C3]
- An independent review committee (IRC) reported a response rate of 53% (95% CI, 44%–62%), with eight patients (6%) achieving a complete response.
- Median duration of response per IRC assessment was 6.7 months (95% CI, 5.6–8.6). Most responses were evident at the first radiological assessment at 6 weeks. However, some patients did not respond until after receiving therapy for more than 6 months.
Dabrafenib
Dabrafenib is an orally available, small-molecule, selective BRAF inhibitor that was approved by the FDA in 2013. It is used for the treatment of patients with unresectable or metastatic melanoma and BRAF V600E pathogenic variants as detected by an FDA-approved test. Dabrafenib and other BRAF inhibitors are not recommended for the treatment of BRAF wild-type melanomas, as in vitro experiments suggest there may be a paradoxical stimulation of MAPK signaling resulting in tumor promotion.
Evidence (dabrafenib):
- An international multicenter trial (BREAK-3 [NCT01227889]) compared dabrafenib with dacarbazine. A total of 250 patients with unresectable stage III or IV melanoma and BRAF V600E pathogenic variants were randomly assigned in a 3:1 ratio to receive dabrafenib (150 mg by mouth daily) or dacarbazine (1,000 mg/m2 IV every 3 weeks). IL-2 was allowed as a previous treatment for advanced disease. The primary end point was PFS; patients could cross over at the time of progressive disease after confirmation by a blinded IRC.[23][Level of evidence B1]
- With 126 events, the HR for PFS was 0.30 (95% CI, 0.18–0.51; P < .0001). The estimated median PFS was 5.1 months for dabrafenib versus 2.7 months for dacarbazine. OS data are limited by the median duration of follow-up and crossover. The partial response rate was 47% and the complete response rate was 3% in patients who received dabrafenib, compared with a partial response rate of 5% and a complete response rate of 2% for those who received dacarbazine.
- The most frequent adverse events in patients treated with dabrafenib were cutaneous findings (i.e., hyperkeratosis, papillomas, palmar-plantar erythrodysesthesia), pyrexia, fatigue, headache, and arthralgia. Cutaneous SCC or keratoacanthoma occurred in 12 patients, basal cell carcinoma occurred in four patients, mycosis fungoides occurred in one patient, and new melanoma occurred in two patients.
MEK inhibitors
Trametinib
Trametinib is an orally available, small-molecule, selective inhibitor of MEK1 and MEK2. BRAF activates MEK1 and MEK2 proteins, which in turn, activate MAPK. Preclinical data suggest that MEK inhibitors can restrain growth and induce cell death of some BRAF-altered human melanoma tumors.
In 2013, the FDA approved trametinib for patients with unresectable or metastatic melanoma with BRAF V600E or V600K pathogenic variants, as determined by an FDA-approved test.
Evidence (trametinib):
- A total of 1,022 patients were screened for BRAF pathogenic variants, resulting in 322 eligible patients (281 with BRAF V600E, 40 with BRAF V600K, and one with both variants).[24] One previous treatment (biological or chemotherapy) was allowed; however, no previous treatment with a BRAF or MEK inhibitor was permitted. Patients were randomly assigned in a 2:1 ratio to receive either trametinib (2 mg every day) or IV chemotherapy (either dacarbazine 1,000 mg/m2 every 3 weeks or paclitaxel 175 mg/m2 every 3 weeks). Crossover for patients randomly assigned to chemotherapy was allowed. The primary end point was PFS.
- The investigator-assessed PFS was 4.8 months in patients who received trametinib and 1.5 months in patients who received chemotherapy (HR for PFS or death, 0.45; 95% CI, 0.33–0.63; P < .001). A radiology review blinded-to-treatment arm resulted in similar outcomes. Median OS has not been reached.
- Adverse events leading to dose interruptions occurred in 35% of patients in the trametinib group and 22% of those in the chemotherapy group. Adverse events leading to dose reductions occurred in 27% of patients who received trametinib and in 10% of those who received chemotherapy.
- The most common adverse events included rash, diarrhea, nausea, vomiting, fatigue, peripheral edema, alopecia, hypertension, and constipation. Cardiomyopathy (7%), interstitial lung disease (2.4%), central serous retinopathy (<1%), and retinal-vein occlusion (<1%) are uncommon but serious adverse events associated with trametinib. On-study cutaneous SCCs were not observed. For more information, visit Fatigue and Nausea and Vomiting Related to Cancer Treatment.
Cobimetinib
Cobimetinib is a small-molecule, selective MEK inhibitor that the FDA approved in 2015 for use in combination with the BRAF inhibitor vemurafenib. For more information, visit the Combination therapy with signal transduction inhibitors section.
KIT inhibitors
Early data suggest that mucosal or acral melanomas with activating pathogenic variants or amplifications in KIT may be sensitive to a variety of c-KIT inhibitors.[25-27] Phase II and phase III trials are available for patients with unresectable stage III or stage IV melanoma and KIT pathogenic variants.
Combination therapy with signal transduction inhibitors
Results from phase III trials comparing three different combinations of BRAF-MEK inhibitors with BRAF inhibitor monotherapy have consistently shown that combination therapy is superior to BRAF monotherapy.
Secondary resistance to BRAF inhibitor monotherapy in patients with BRAF V600 pathogenic variants may be associated with reactivation of the MAPK pathway. Therefore, combinations of signal transduction inhibitors that block different sites in the same pathway or sites in multiple pathways are an active area of research.
BRAF inhibitor plus MEK inhibitors
Dabrafenib plus trametinib
Evidence (dabrafenib plus trametinib):
- Previously untreated. An international, double-blind, phase III trial (COMBI-d [NCT01584648]) without crossover included 423 previously untreated patients with unresectable stage IIIC or stage IV melanoma and BRAF V600E or V600K pathogenic variants. Patients were randomly assigned to receive either the combination of dabrafenib (150 mg by mouth twice a day) plus trametinib (2 mg by mouth every day) or dabrafenib plus placebo. The primary end point was investigator-assessed PFS. The protocol included a prespecified interim analysis for OS at the time of analysis of the primary end point. Patients were stratified by baseline LDH levels and BRAF genotype.[28][Level of evidence B1]
- Median PFS was 9.3 months for the combination versus 8.8 months for dabrafenib plus placebo. The HRdeath or progression was 0.75 (95% CI, 0.57–0.99; P = .03). Updated data at the time of final analysis of OS revealed a median PFS of 11.0 months for the combination versus 8.8 months for dabrafenib plus placebo. The HRPFS or death was 0.67 (95% CI, 0.53–0.84; P = .0004; unadjusted for multiple testing).[29]
- A prespecified final analysis of OS was conducted at 70% of events. Median OS was 25.1 months in the dabrafenib-plus-trametinib group (66% of events) versus 18.7 months in the dabrafenib-plus-placebo group (76% of events). The HR was 0.71 (95% CI, 0.55–0.92; P = 0.01).
- Permanent discontinuations of study drugs were reported in 9% of patients who received the combination and in 5% of patients treated with dabrafenib only.
- The incidence of grade 3 to grade 4 adverse events was similar between the groups (35% with the combination and 37% with dabrafenib only). Pyrexia occurred more frequently with the combination and was treated with immediate temporary cessation of the study drug in either group; prophylactic glucocorticoids may prevent recurring episodes. Hyperproliferative cutaneous events, including cutaneous SCCs, were considered related to paradoxical activation of the MAPK pathway and occurred less frequently with the addition of the MEK inhibitor. Rare, but serious, adverse events included decreased ejection fraction and chorioretinopathy.
- Previously untreated. An international, open-label, phase III trial (COMBI-v [NCT01597908]) included 704 previously untreated patients with metastatic melanoma and BRAF V600 pathogenic variants. Patients were randomly assigned to receive standard doses of either the combination of dabrafenib plus trametinib or vemurafenib as first-line therapy. The primary end point was OS.[30][Level of evidence A1]
- An interim analysis for OS was planned when 202 of the final 288 events occurred. Per protocol, the DMSB used adjusted efficacy boundaries for actual events (222) (2-sided P < .0214 for efficacy and P > .2210 for futility). The DMSB recommended stopping for efficacy, and the interim analysis is considered to be the final analysis of OS. A protocol amendment was issued to allow crossover to the combination therapy arm.
- A total of 100 patients (28%) in the combination arm and 122 (35%) in the vemurafenib group had died (HR, 0.69; 95% CI, 0.53–0.89; P = .005). Median OS for patients treated with vemurafenib was 17.2 months; the median has not been reached in the combination therapy arm.
- Previously untreated. A pooled analysis of the 563 patients who were randomly assigned to receive dabrafenib plus trametinib in the COMBI-d (double-blinded) and COMBI-v (open label) trials (described above) provides estimated 5-year outcomes.[31] After the study, 53% of patients received subsequent treatment; two-thirds of these patients received immunotherapy.
- Sixty-three percent of patients died (64% in COMBI-d and 61% in COMBI-v) after a median follow-up of 22 months (range, 0−76 months).
- The estimated 5-year OS rate was 34% (95% CI, 30%−38%), and the investigator-assessed PFS rate was 19% (95% CI, 15%−22%).
- A complete response, which occurred in 109 patients (19%), was associated with an OS rate of 71% (95% CI, 62%−79%) at 5 years.
Vemurafenib plus cobimetinib
Evidence (vemurafenib plus cobimetinib):
- Previously untreated. An international phase III trial included 495 patients with previously untreated, unresectable, stage IIIC or stage IV melanoma and BRAF V600 pathogenic variants. Patients were randomly assigned to receive either the combination of vemurafenib (960 mg by mouth every day) and cobimetinib (60 mg by mouth every day for 21 days followed by a 7-day rest period) or vemurafenib plus placebo. The primary end point was investigator-assessed PFS. Crossover at time of PFS was not allowed. Patients were stratified by stage and geographic region. Two interim analyses of OS were prespecified, with the first specified at the time of analysis of the primary end point.[32][Level of evidence B1]
- The median PFS was 9.9 months in patients who received the combination versus 6.2 months in patients treated with vemurafenib plus placebo. The HRprogression or death was 0.51 (95% CI, 0.39–0.68; P = .001).
- The first interim analysis of OS is immature because of the few events in both arms; therefore, median survival was not reached in either study group.
- Rate of withdrawal of therapy caused by adverse events was similar between the groups (13% for patients treated with the combination and 12% for patients treated with vemurafenib only). Six deaths were attributed to adverse events in the combination group, and three deaths were attributed to adverse events in the vemurafenib-only group.
- The incidence of grade 3 to grade 4 adverse events was similar between the groups (62% in patients treated with the combination and 58% in patients treated with vemurafenib alone). Rare, but serious, adverse events included chorioretinopathy, retinal detachment, decreased ejection fraction, and QT prolongation. Hyperproliferative cutaneous events, including cutaneous SCC, were considered to be related to paradoxical activation of the MAPK pathway and occurred less frequently with the addition of the MEK inhibitor.
Encorafenib plus binimetinib
Encorafenib is a small-molecule BRAF inhibitor, and binimetinib is a small-molecule MEK inhibitor. The combination of these two agents is approved for the treatment of unresectable or metastatic melanoma with BRAF V600E or V600K pathogenic variants, as detected by an FDA-approved test. The combination has demonstrated improved PFS and OS compared with vemurafenib. However, neither is approved as single-agent therapy.
Evidence (encorafenib plus binimetinib):
- Previously untreated or progression on or after first-line immunotherapy. An international, open-label, phase III trial (COLUMBUS [NCT01909453]) included 577 patients with stage IIIB, IIIC, or IV melanoma and BRAF V600 pathogenic variants. Patients were randomly assigned in a 1:1:1 ratio to receive encorafenib (450 mg every day) plus binimetinib (45 mg twice a day), encorafenib monotherapy (300 mg every day), or vemurafenib monotherapy (960 mg twice a day).[33,34] The primary end point was PFS for the combination versus vemurafenib alone as assessed by a blinded IRC with a secondary end point of OS.
- Approximately 5% of patients had received previous checkpoint inhibitor therapy.
- With a median follow-up of 16.6 months, the median PFS was 14.9 months (95% CI, 11.0–18.5) with the combination versus 7.3 months (95% CI, 5.6–8.2) with vemurafenib alone. The HRprogression or death was 0.54 (95% CI, 0.41–0.71; 2-sided P < .0001).
- With a median follow-up of 36.8 months for the secondary end point of OS, the median OS was 33.6 months (95% CI, 24.4–39.2) for patients in the combination arm and 16.9 months (95% CI, 14.0–24.5) for patients treated with vemurafenib alone (HR, 0.61; 95% CI, 0.47–0.79; P < .0001). Subsequent treatments after discontinuation of the study drug were received by 42% of patients in the combination group and 62% of patients in the vemurafenib-alone group.[33,34][Level of evidence A1]
- The incidence of grade 3 to 4 adverse events was 58% with combination therapy and 63% with vemurafenib alone. Serious adverse events occurred in 34% of the combination group and 37% of the vemurafenib-alone group. The most common adverse events in the combination group included gastrointestinal symptoms and elevation of gamma-glutamyl transferase (GGT), elevation of creatine phosphokinase (CPK), left ventricular dysfunction (8%), and serous retinopathy (20%), mostly grade 1 to 2 (monitoring guidelines are provided in the drug label). Patients who received vemurafenib had more pyrexia and cutaneous toxicities. Study drug discontinuations from adverse events occurred in 15% of patients in the combination group and 17% of patients in the vemurafenib-alone group. No deaths were related to treatment; however, one death from suicide occurred in the combination arm.
Combination signal transduction inhibitor therapy plus PD-L1 inhibitor
Cobimetinib and vemurafenib plus atezolizumab
Evidence (cobimetinib and vemurafenib plus atezolizumab):
- A double-blind, placebo-controlled, multicenter trial (IMspire150 [NCT02908672]) included 514 patients with unresectable stage IIIC or metastatic melanoma and BRAF V600 pathogenic variants. Patients were randomly assigned (1:1) to receive first-line therapy with cobimetinib plus vemurafenib with either atezolizumab or placebo.[35] Eligibility criteria included ECOG PS scores of 0 to 1, measurable disease, and no previous systemic treatment for metastatic melanoma. Patients with untreated or actively progressing brain metastases or a history of serious autoimmune disease were excluded. Previous adjuvant therapy was allowed (14% of patients).
After all patients in both arms received a 28-day cycle of cobimetinib and vemurafenib, patients received atezolizumab (840 mg IV every 2 weeks) or placebo in addition to the combination BRAF-MEK inhibitor therapy. The primary efficacy end point was investigator-assessed PFS per RECIST 1.1 criteria.
- At a median follow-up of 19 months, the primary investigator median PFS was 15 months (95% CI, 11.4−18.4) in the atezolizumab arm and 11 months (95% CI, 9.3−12.7) in the placebo arm (HR, 0.78; 95% CI, 0.63−0.97; P = .0249).
- An IRC assessment of the triplet therapy found a PFS of 16 months (95% CI, 11.3−18.5), compared with 12 months in the control arm (95% CI, 10.8−14.7) (HR, 0.85; 95% CI, 0.67−1.07).
- Objective response rates and complete responses were similar between the treatment groups.
- Data are not mature for OS.
- Serious adverse events and treatment discontinuations because of toxicity were similar between the arms. Grade 5 adverse events occurred in seven patients in each arm. Two patients with hepatic failure in the atezolizumab group and one patient with pulmonary hemorrhage in the control group were considered treatment related.
The impact of triplet therapy on OS, or when compared with checkpoint inhibitor monotherapy, or to sequential therapy with combination BRAF-MEK inhibitor therapy, preceded or followed by checkpoint inhibition (ongoing trial NCT02224781) is unknown.
Tumor-infiltrating lymphocyte (TIL) therapy
Lifileucel
Lifileucel is an autologous TIL therapy. The FDA approved lifileucel for the treatment of adult patients with unresectable or metastatic melanoma who have previously received a PD-1 blocking antibody and, if BRAF V600 variant–positive, a BRAF inhibitor with or without a MEK inhibitor.
Evidence (lifileucel):
- A multicenter, multicohort, open-label trial (C-144-01 [NCT02360579]) evaluated the efficacy and safety of lifileucel in patients with unresectable or metastatic melanoma. The pivotal cohort (cohort 4) included 73 adult patients who had received prior anti–PD-1 therapy and, if BRAF V600 variant–positive, a BRAF inhibitor with or without a MEK inhibitor. Most patients had also received prior ipilimumab. All patients had an ECOG performance status of 0 or 1.
Patients underwent nonmyeloablative lymphodepleting chemotherapy followed by a single infusion of lifileucel and subsequent high-dose IL-2. Tumors were resected and processed to manufacture the autologous TIL product prior to lymphodepletion. Efficacy was assessed according to objective response rate and duration of response.[36][Level of evidence B4]
- At a median follow-up of 27.6 months. the objective response rate was 31.5% (95% CI, 21.1%–43.4%), including 4 complete responses and 19 partial responses. The median duration of response was not reached, and 44% of responses lasted 12 months or longer.[36]
- In a supporting pooled efficacy set that included 153 patients, the objective response rate was 31.4% (95% CI, 24.1%–39.4%). The median duration of response was not reached, with 54.2% of responses maintained at 12 months.
- In a subsequent analysis of the pooled efficacy cohort at a median follow-up of 48.1 months, the median OS was 13.9 months. The 3-year and 4-year OS rates were 28.4% and 21.9%, respectively.[37]
- The most common grade 3 or higher adverse events were associated with lymphodepleting chemotherapy and IL-2 administration, including thrombocytopenia, anemia, febrile neutropenia, and hypotension.
- Treatment-related mortality was not observed, although high-grade toxicities required inpatient monitoring and supportive care. This is an intense regimen that may only be offered at tertiary centers with experience in its administration.
Lifileucel is given as a one-time treatment and is the first individualized TIL therapy approved for patients with advanced melanoma refractory to PD-1 blockade.
Intralesional therapy
Talimogene laherparepvec (T-VEC)
T-VEC is a genetically modified, herpes simplex virus type 1 (HSV1) oncolytic therapy approved for local intralesional injection into unresectable cutaneous, subcutaneous, and nodal lesions in patients with melanoma that recurs after initial surgery. T-VEC is designed to replicate within tumors, causing lysis, and to produce granulocyte-macrophage colony-stimulating factor (GM-CSF). Release of antigens together with virally derived GM-CSF may promote an antitumor immune response. However, the exact mechanism of action is unknown.
The approval of T-VEC by the FDA was based on data that demonstrated shrinkage of lesions. However, improvement of OS, an effect on visceral metastases, or improvement in quality of life has not been shown.
Evidence (T-VEC):
- In a multinational open-label trial (NCT00769704), 436 patients were randomly assigned (2:1) to receive either intralesional T-VEC or subcutaneous GM-CSF for at least 6 months or until there were no more injectable lesions.[38][Level of evidence B3] Eligible patients had stage IIIB, IIIC, or IV melanoma with unresectable, bidimensionally measurable lesions. The primary end point was durable response rate (DRR) (complete response or partial response lasting for >6 months) as assessed by independent review. The study was stratified by site of first recurrence, presence of liver metastases, disease stage, and previous nonadjuvant systemic treatment.
- The median patient age was 63 years (range, 22–94), 70% of patients had a baseline ECOG PS score of 0, 30% had stage III disease, and 70% had stage IV disease (27% M1a; 21% M1b; and 22% M1c). Previous therapy for melanoma had been received by 53% of the patients.
- The first dose only was administered at 106 plaque-forming units (pfu)/mL to a maximum of 4 mL for all lesions combined. Subsequent doses were administered at 8 pfu/mL up to 4.0 mL for all injected lesions combined with the injected volume based on the size of the lesion. Injection into visceral lesions was not allowed.
- In patients treated with T-VEC, 16% (95% CI, 12.0%–20.5%) had a DRR versus 2% (95% CI, 0%–4.5%) in patients who received GM-CSF. Subgroup analysis suggested that the differences in DRRs between T-VEC versus GM-CSF may be greater in earlier-stage disease and treatment-naïve disease, as follows:
- In patients with stage IIIB and IIIC disease, the DRR was 33% in the T-VEC group and 0% in the GM-CSF group.
- In patients with stage IV M1a disease, the DRR was 16% in the T-VEC group and 2% in the GM-CSF group.
- In patients with stage IV M1b disease, the DRR was 3% in the T-VEC group and 4% in the GM-CSF group.
- In patients with stage IV M1c disease, the DRR was 7% in the T-VEC group and 3% in the GM-CSF group.
- Patients treated with T-VEC or GM-CSF as first-line therapy had a DRR of 24% versus 0%, respectively. However, patients who received treatment as second-line therapy or greater had a DRR of 10% versus 4%, respectively.
- The median duration of exposure to T-VEC was 23 weeks (5.3 months), with 26 patients exposed for more than 1 year. The most common adverse events in the T-VEC group were fatigue (50%), chills (49%), pyrexia (43%), nausea (36%), influenza-like illness (30%), and injection site pain. The rate of discontinuation resulting from toxicity to T-VEC was 4%, versus 2% in the GM-CSF group. Of the ten deaths in patients treated with T-VEC, eight deaths were considered the result of PD-1, salmonella infection, and one myocardial infarction; none were considered related to therapy, based on findings of the investigator.
Precautions: T-VEC is a live attenuated HSV and may cause life-threatening, disseminated herpetic infection. It is contraindicated in immunocompromised or pregnant patients. Health care providers and close contacts should avoid direct contact with injected lesions. Biohazard precautions for preparation, administration, and handling are provided in the label.
Detailed prescribing information by treatment cycle and lesion size are provided in the FDA label.
Palliative therapy
Regional lymphadenectomy may be used as palliative care for melanoma that is metastatic to distant, lymph node–bearing areas. Resection may be used as palliative care for isolated metastases to the lung, gastrointestinal tract, bone, or sometimes the brain, with occasional long-term survival.[39-41]
Chemotherapy
Dacarbazine was approved in 1970 based on objective response rates. Phase III trials indicate an objective response rate of 10% to 20%, with rare complete responses observed. An impact on OS has not been demonstrated in randomized trials.[6,21,42-44] When used as a control arm for recent registration trials of ipilimumab and vemurafenib in previously untreated patients with metastatic melanoma, dacarbazine was inferior for OS.
Temozolomide, an oral alkylating agent that hydrolyzes to the same active moiety as dacarbazine, appeared to be similar to dacarbazine (IV administration) in a randomized phase III trial with a primary end point of OS. However, the trial was designed for superiority, and the sample size was inadequate to prove equivalency.[43]
The objective response rate to dacarbazine and the nitrosoureas, carmustine and lomustine, is approximately 10% to 20%.[42,45-47] Responses are usually short-lived, ranging from 3 to 6 months, although long-term remissions can occur in a limited number of patients who attain a complete response.[45,47]
A randomized trial compared IV dacarbazine with temozolomide, an oral agent. OS was 6.4 months for dacarbazine versus 7.7 months for temozolomide (HR, 1.18; 95% CI, 0.92–1.52). While these data suggested similarity between dacarbazine and temozolomide, no benefit in survival has been demonstrated for either dacarbazine or temozolomide. Therefore, evidence of similarity did not result in FDA approval of temozolomide.[43][Level of evidence A1]
An extended schedule and escalated dose of temozolomide was compared with dacarbazine in a multicenter trial by the European Organisation for Research and Treatment of Cancer (EORTC) (EORTC-18032 [NCT00101218]) that randomly assigned 859 patients. No improvement was seen in OS or PFS for the temozolomide group, and this dose and schedule resulted in more toxicity than standard-dose, single-agent dacarbazine.[48][Level of evidence A1]
Two randomized phase III trials in previously untreated patients with metastatic melanoma (resulting in FDA approval for vemurafenib [21] and ipilimumab [6]) included dacarbazine as the standard therapy arm. Both vemurafenib (in BRAF V600–altered melanoma) and ipilimumab showed superior OS compared with dacarbazine in the two separate trials.
Other agents with modest, single-agent activity include vinca alkaloids, platinum compounds, and taxanes.[45,46]
Attempts to develop combination regimens that incorporate chemotherapy (e.g., multiagent chemotherapy,[49,50] combinations of chemotherapy and tamoxifen,[51-53] and combinations of chemotherapy and immunotherapy [9,10,39-41,49,54]) have not demonstrated an improvement in OS.
A published data meta-analysis of 18 randomized trials (15 of which had survival information) that compared chemotherapy with biochemotherapy (i.e., the same chemotherapy plus interferon alone or with IL-2) reported no impact on OS.[55][Level of evidence A1]
Radiation therapy
Although melanoma is a relatively radiation-resistant tumor, palliative radiation therapy may alleviate symptoms. Retrospective studies have shown that symptom relief and some shrinkage of the tumor with radiation therapy may occur in patients with:[56,57]
- Multiple brain metastases.
- Bone metastases.
- Spinal cord compression.
The most effective dose-fractionation schedule for palliation of melanoma metastatic to the bone or spinal cord is unclear, but high-dose-per-fraction schedules are sometimes used to overcome tumor resistance. For more information, visit Cancer Pain.
Current Clinical Trials
Use our advanced clinical trial search to find NCI-supported cancer clinical trials that are now enrolling patients. The search can be narrowed by location of the trial, type of treatment, name of the drug, and other criteria. General information about clinical trials is also available.
References
- Robert C, Ribas A, Wolchok JD, et al.: Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet 384 (9948): 1109-17, 2014. [PUBMED Abstract]
- Robert C, Schachter J, Long GV, et al.: Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med 372 (26): 2521-32, 2015. [PUBMED Abstract]
- Schachter J, Ribas A, Long GV, et al.: Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 390 (10105): 1853-1862, 2017. [PUBMED Abstract]
- Robert C, Long GV, Brady B, et al.: Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 372 (4): 320-30, 2015. [PUBMED Abstract]
- Zhao X, Suryawanshi S, Hruska M, et al.: Assessment of nivolumab benefit-risk profile of a 240-mg flat dose relative to a 3-mg/kg dosing regimen in patients with advanced tumors. Ann Oncol 28 (8): 2002-2008, 2017. [PUBMED Abstract]
- Robert C, Thomas L, Bondarenko I, et al.: Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 364 (26): 2517-26, 2011. [PUBMED Abstract]
- Hodi FS, O'Day SJ, McDermott DF, et al.: Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363 (8): 711-23, 2010. [PUBMED Abstract]
- Yervoy (Ipilimumab): Immune-Mediated Adverse Reaction Management Guide. Princeton, NJ: Bristol-Myers Squibb, 2011. Available online. Last accessed May 1, 2025.
- Atkins MB, Lotze MT, Dutcher JP, et al.: High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol 17 (7): 2105-16, 1999. [PUBMED Abstract]
- Atkins MB, Kunkel L, Sznol M, et al.: High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J Sci Am 6 (Suppl 1): S11-4, 2000. [PUBMED Abstract]
- Postow MA, Chesney J, Pavlick AC, et al.: Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 372 (21): 2006-17, 2015. [PUBMED Abstract]
- Larkin J, Chiarion-Sileni V, Gonzalez R, et al.: Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 373 (1): 23-34, 2015. [PUBMED Abstract]
- Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al.: Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med 377 (14): 1345-1356, 2017. [PUBMED Abstract]
- Wolchok JD, Chiarion-Sileni V, Rutkowski P, et al.: Final, 10-Year Outcomes with Nivolumab plus Ipilimumab in Advanced Melanoma. N Engl J Med 392 (1): 11-22, 2025. [PUBMED Abstract]
- Tawbi HA, Forsyth PA, Algazi A, et al.: Combined Nivolumab and Ipilimumab in Melanoma Metastatic to the Brain. N Engl J Med 379 (8): 722-730, 2018. [PUBMED Abstract]
- Tawbi HA, Schadendorf D, Lipson EJ, et al.: Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N Engl J Med 386 (1): 24-34, 2022. [PUBMED Abstract]
- Heidorn SJ, Milagre C, Whittaker S, et al.: Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140 (2): 209-21, 2010. [PUBMED Abstract]
- Hatzivassiliou G, Song K, Yen I, et al.: RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464 (7287): 431-5, 2010. [PUBMED Abstract]
- Poulikakos PI, Zhang C, Bollag G, et al.: RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464 (7287): 427-30, 2010. [PUBMED Abstract]
- Su F, Viros A, Milagre C, et al.: RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 366 (3): 207-15, 2012. [PUBMED Abstract]
- Chapman PB, Hauschild A, Robert C, et al.: Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364 (26): 2507-16, 2011. [PUBMED Abstract]
- Sosman JA, Kim KB, Schuchter L, et al.: Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 366 (8): 707-14, 2012. [PUBMED Abstract]
- Hauschild A, Grob JJ, Demidov LV, et al.: Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380 (9839): 358-65, 2012. [PUBMED Abstract]
- Flaherty KT, Robert C, Hersey P, et al.: Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367 (2): 107-14, 2012. [PUBMED Abstract]
- Hodi FS, Friedlander P, Corless CL, et al.: Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol 26 (12): 2046-51, 2008. [PUBMED Abstract]
- Guo J, Si L, Kong Y, et al.: Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol 29 (21): 2904-9, 2011. [PUBMED Abstract]
- Carvajal RD, Antonescu CR, Wolchok JD, et al.: KIT as a therapeutic target in metastatic melanoma. JAMA 305 (22): 2327-34, 2011. [PUBMED Abstract]
- Long GV, Stroyakovskiy D, Gogas H, et al.: Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 371 (20): 1877-88, 2014. [PUBMED Abstract]
- Long GV, Stroyakovskiy D, Gogas H, et al.: Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet 386 (9992): 444-51, 2015. [PUBMED Abstract]
- Robert C, Karaszewska B, Schachter J, et al.: Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 372 (1): 30-9, 2015. [PUBMED Abstract]
- Robert C, Grob JJ, Stroyakovskiy D, et al.: Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N Engl J Med 381 (7): 626-636, 2019. [PUBMED Abstract]
- Larkin J, Ascierto PA, Dréno B, et al.: Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 371 (20): 1867-76, 2014. [PUBMED Abstract]
- Dummer R, Ascierto PA, Gogas HJ, et al.: Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 19 (5): 603-615, 2018. [PUBMED Abstract]
- Dummer R, Ascierto PA, Gogas HJ, et al.: Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 19 (10): 1315-1327, 2018. [PUBMED Abstract]
- Gutzmer R, Stroyakovskiy D, Gogas H, et al.: Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAFV600 mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 395 (10240): 1835-1844, 2020. [PUBMED Abstract]
- Chesney J, Lewis KD, Kluger H, et al.: Efficacy and safety of lifileucel, a one-time autologous tumor-infiltrating lymphocyte (TIL) cell therapy, in patients with advanced melanoma after progression on immune checkpoint inhibitors and targeted therapies: pooled analysis of consecutive cohorts of the C-144-01 study. J Immunother Cancer 10 (12): , 2022. [PUBMED Abstract]
- Medina T, Chesney JA, Whitman E, et al.: Long-term efficacy and safety of lifileucel tumor-infiltrating lymphocyte (TIL) cell therapy in patients with advanced melanoma: a 4-year analysis of the C-144–01 study. [Abstract] J Immunother Cancer 11 (Suppl 1): A-776, 873, 2023.
- Andtbacka RH, Kaufman HL, Collichio F, et al.: Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J Clin Oncol 33 (25): 2780-8, 2015. [PUBMED Abstract]
- Leo F, Cagini L, Rocmans P, et al.: Lung metastases from melanoma: when is surgical treatment warranted? Br J Cancer 83 (5): 569-72, 2000. [PUBMED Abstract]
- Ollila DW, Hsueh EC, Stern SL, et al.: Metastasectomy for recurrent stage IV melanoma. J Surg Oncol 71 (4): 209-13, 1999. [PUBMED Abstract]
- Gutman H, Hess KR, Kokotsakis JA, et al.: Surgery for abdominal metastases of cutaneous melanoma. World J Surg 25 (6): 750-8, 2001. [PUBMED Abstract]
- Chapman PB, Einhorn LH, Meyers ML, et al.: Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with metastatic melanoma. J Clin Oncol 17 (9): 2745-51, 1999. [PUBMED Abstract]
- Middleton MR, Grob JJ, Aaronson N, et al.: Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol 18 (1): 158-66, 2000. [PUBMED Abstract]
- Avril MF, Aamdal S, Grob JJ, et al.: Fotemustine compared with dacarbazine in patients with disseminated malignant melanoma: a phase III study. J Clin Oncol 22 (6): 1118-25, 2004. [PUBMED Abstract]
- Anderson CM, Buzaid AC, Legha SS: Systemic treatments for advanced cutaneous melanoma. Oncology (Huntingt) 9 (11): 1149-58; discussion 1163-4, 1167-8, 1995. [PUBMED Abstract]
- Wagner JD, Gordon MS, Chuang TY, et al.: Current therapy of cutaneous melanoma. Plast Reconstr Surg 105 (5): 1774-99; quiz 1800-1, 2000. [PUBMED Abstract]
- Mays SR, Nelson BR: Current therapy of cutaneous melanoma. Cutis 63 (5): 293-8, 1999. [PUBMED Abstract]
- Patel PM, Suciu S, Mortier L, et al.: Extended schedule, escalated dose temozolomide versus dacarbazine in stage IV melanoma: final results of a randomised phase III study (EORTC 18032). Eur J Cancer 47 (10): 1476-83, 2011. [PUBMED Abstract]
- Kirkwood JM, Strawderman MH, Ernstoff MS, et al.: Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J Clin Oncol 14 (1): 7-17, 1996. [PUBMED Abstract]
- Kirkwood JM, Ibrahim JG, Sondak VK, et al.: High- and low-dose interferon alfa-2b in high-risk melanoma: first analysis of intergroup trial E1690/S9111/C9190. J Clin Oncol 18 (12): 2444-58, 2000. [PUBMED Abstract]
- Kirkwood JM, Ibrahim J, Lawson DH, et al.: High-dose interferon alfa-2b does not diminish antibody response to GM2 vaccination in patients with resected melanoma: results of the Multicenter Eastern Cooperative Oncology Group Phase II Trial E2696. J Clin Oncol 19 (5): 1430-6, 2001. [PUBMED Abstract]
- Hancock BW, Wheatley K, Harris S, et al.: Adjuvant interferon in high-risk melanoma: the AIM HIGH Study--United Kingdom Coordinating Committee on Cancer Research randomized study of adjuvant low-dose extended-duration interferon Alfa-2a in high-risk resected malignant melanoma. J Clin Oncol 22 (1): 53-61, 2004. [PUBMED Abstract]
- Koops HS, Vaglini M, Suciu S, et al.: Prophylactic isolated limb perfusion for localized, high-risk limb melanoma: results of a multicenter randomized phase III trial. European Organization for Research and Treatment of Cancer Malignant Melanoma Cooperative Group Protocol 18832, the World Health Organization Melanoma Program Trial 15, and the North American Perfusion Group Southwest Oncology Group-8593. J Clin Oncol 16 (9): 2906-12, 1998. [PUBMED Abstract]
- Lee ML, Tomsu K, Von Eschen KB: Duration of survival for disseminated malignant melanoma: results of a meta-analysis. Melanoma Res 10 (1): 81-92, 2000. [PUBMED Abstract]
- Ives NJ, Stowe RL, Lorigan P, et al.: Chemotherapy compared with biochemotherapy for the treatment of metastatic melanoma: a meta-analysis of 18 trials involving 2,621 patients. J Clin Oncol 25 (34): 5426-34, 2007. [PUBMED Abstract]
- Rate WR, Solin LJ, Turrisi AT: Palliative radiotherapy for metastatic malignant melanoma: brain metastases, bone metastases, and spinal cord compression. Int J Radiat Oncol Biol Phys 15 (4): 859-64, 1988. [PUBMED Abstract]
- Herbert SH, Solin LJ, Rate WR, et al.: The effect of palliative radiation therapy on epidural compression due to metastatic malignant melanoma. Cancer 67 (10): 2472-6, 1991. [PUBMED Abstract]
Latest Updates to This Summary (05/02/2025)
The PDQ cancer information summaries are reviewed regularly and updated as new information becomes available. This section describes the latest changes made to this summary as of the date above.
Treatment of Stage II Melanoma
Revised text about the results of a trial that randomly assigned patients with completely resected stage IIB or stage IIC melanoma to receive either pembrolizumab or placebo (cited Luke et al. as reference 19).
Treatment of Resectable Stage III Melanoma
Revised text about the final analysis results of a multinational double-blind trial that randomly assigned patients with stage IIIA, IIIB, or IIIC melanoma with BRAF V600E or V600K pathogenic variants who underwent completion lymphadenectomy to receive either dabrafenib plus trametinib or two matched placebo tablets (cited Long et al. as reference 32).
Treatment of Unresectable Stage III, Stage IV, and Recurrent Melanoma
Revised the list of treatment options for unresectable stage III, stage IV, and recurrent melanoma to include tumor-infiltrating lymphocyte therapy.
Revised text about the final analysis results of an international double-blind trial that randomly assigned 945 previously untreated patients with unresectable stage III or IV melanoma to receive nivolumab plus placebo, nivolumab plus ipilimumab followed by nivolumab, or ipilimumab plus placebo (cited Wolchok et al. as reference 14).
Added Tumor-infiltrating lymphocyte therapy as a new subsection.
This summary is written and maintained by the PDQ Adult Treatment Editorial Board, which is editorially independent of NCI. The summary reflects an independent review of the literature and does not represent a policy statement of NCI or NIH. More information about summary policies and the role of the PDQ Editorial Boards in maintaining the PDQ summaries can be found on the About This PDQ Summary and PDQ® Cancer Information for Health Professionals pages.
About This PDQ Summary
Purpose of This Summary
This PDQ cancer information summary for health professionals provides comprehensive, peer-reviewed, evidence-based information about the treatment of melanoma. It is intended as a resource to inform and assist clinicians in the care of their patients. It does not provide formal guidelines or recommendations for making health care decisions.
Reviewers and Updates
This summary is reviewed regularly and updated as necessary by the PDQ Adult Treatment Editorial Board, which is editorially independent of the National Cancer Institute (NCI). The summary reflects an independent review of the literature and does not represent a policy statement of NCI or the National Institutes of Health (NIH).
Board members review recently published articles each month to determine whether an article should:
- be discussed at a meeting,
- be cited with text, or
- replace or update an existing article that is already cited.
Changes to the summaries are made through a consensus process in which Board members evaluate the strength of the evidence in the published articles and determine how the article should be included in the summary.
The lead reviewer for Melanoma Treatment is:
- Shaheer A. Khan, DO (Columbia University Irving Medical Center)
Any comments or questions about the summary content should be submitted to Cancer.gov through the NCI website's Email Us. Do not contact the individual Board Members with questions or comments about the summaries. Board members will not respond to individual inquiries.
Levels of Evidence
Some of the reference citations in this summary are accompanied by a level-of-evidence designation. These designations are intended to help readers assess the strength of the evidence supporting the use of specific interventions or approaches. The PDQ Adult Treatment Editorial Board uses a formal evidence ranking system in developing its level-of-evidence designations.
Permission to Use This Summary
PDQ is a registered trademark. Although the content of PDQ documents can be used freely as text, it cannot be identified as an NCI PDQ cancer information summary unless it is presented in its entirety and is regularly updated. However, an author would be permitted to write a sentence such as “NCI’s PDQ cancer information summary about breast cancer prevention states the risks succinctly: [include excerpt from the summary].”
The preferred citation for this PDQ summary is:
PDQ® Adult Treatment Editorial Board. PDQ Melanoma Treatment. Bethesda, MD: National Cancer Institute. Updated <MM/DD/YYYY>. Available at: https://www.cancer.gov/types/skin/hp/melanoma-treatment-pdq. Accessed <MM/DD/YYYY>. [PMID: 26389469]
Images in this summary are used with permission of the author(s), artist, and/or publisher for use within the PDQ summaries only. Permission to use images outside the context of PDQ information must be obtained from the owner(s) and cannot be granted by the National Cancer Institute. Information about using the illustrations in this summary, along with many other cancer-related images, is available in Visuals Online, a collection of over 2,000 scientific images.
Disclaimer
Based on the strength of the available evidence, treatment options may be described as either “standard” or “under clinical evaluation.” These classifications should not be used as a basis for insurance reimbursement determinations. More information on insurance coverage is available on Cancer.gov on the Managing Cancer Care page.
Contact Us
More information about contacting us or receiving help with the Cancer.gov website can be found on our Contact Us for Help page. Questions can also be submitted to Cancer.gov through the website’s Email Us.