Stream the Episode
Find this episode on YouTube:
Additional Information
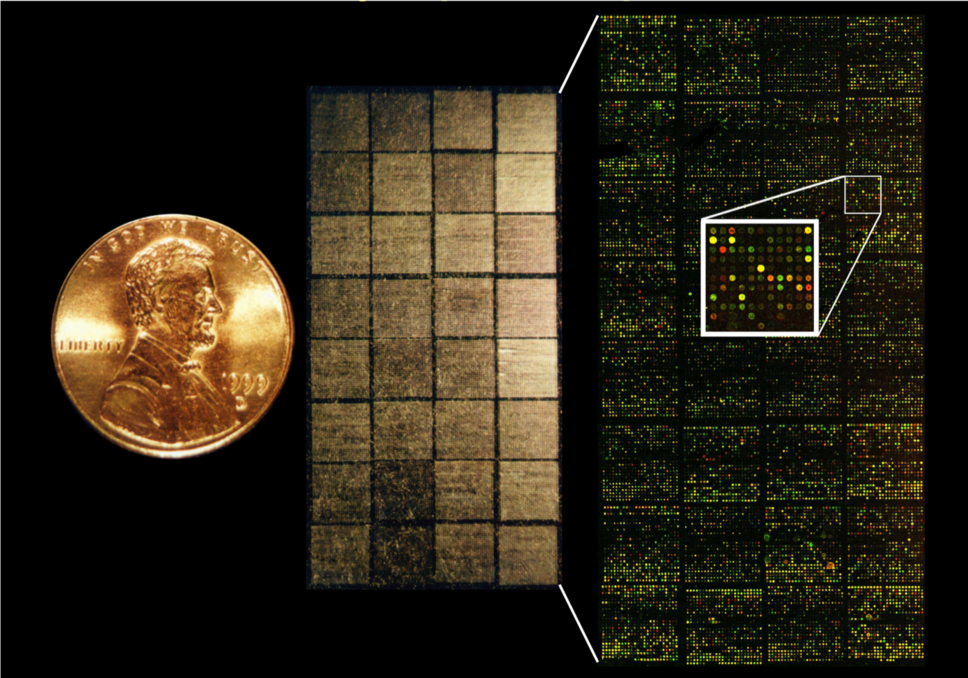
Ash and Lou's study of diffuse large B-cell lymphoma using their Lymphochip was published in Nature
Music used on this episode is by Loyalty Freak Music.
Episode Transcript
PEGGY WANG: Research in cancer genomics—it's hard! When you're trying to figure out a deadly complex disease or change how patients are treated, your job is kind of hard by design. But there's a lot more to it that we don't often talk about. If you're a researcher, other people in the same field, maybe even in your own lab, don't know all of what you go through. What people do know is usually a scientific paper, which shows the work as a seamless, linear process from one question to one conclusion. In this podcast, we want to break down this façade and show research as it is: an organic, messy process, often from humble beginnings.
[SONG: "Friend to Friend"]
LOU STAUDT: And it just sort of dawned on me that here I was in the National Cancer Institute, maybe I should consider working on cancer. So I just went to the NIH library and got out the textbooks on lymphoma and started reading them. And they looked like Greek to me.
PEGGY: This is Dr. Lou Staudt.
LOU: So I split my time between running large genomic programs through the Center for Cancer Genomics for the Cancer Institute and trying to cure lymphoma.
PEGGY: Today, genomics is pretty much at the core of all Lou's work. But when Lou first started his research, he didn't know anything about genomics, and little about cancer. This is the story about how all of that changed. I'm Peggy Wang and this is Personal Genomics: a podcast from CCG--the Center for Cancer Genomics at the National Cancer Institute. Our first story is from CCG's own director, Lou Staudt.
LOU: When I first came here I was just a dyed-in-the-wool basic scientist.
PEGGY: Lou came to NCI in '88 and started out working out on basic biology problems. He had both medical training and research training, but for the most part, those halves of his training remained separate, even though he has always wanted…
LOU: To do something that was really related to patients, even though I had decided long ago not to actually do patient care.
PEGGY: And one day Lou realized, he might be able to achieve this by working on cancer.
LOU: So I had spent my entire life working on B lymphocytes.
PEGGY: A type of white blood cell.
LOU: So I said, well, what is a cancer of a B lymphocyte? What would that look like? Well, that's called a lymphoma.
PEGGY: So Lou went to the library, pulled out some books about lymphoma, and had a lot of trouble understanding them.
LOU: These textbooks, these seemed to be just sort of broad descriptions based on how the lymphomas looked to the pathologist under the microscope. So maybe we could use the tools of molecular biology to find subgroups of patients that were biologically different. And maybe if we were lucky, also find that they had different responses to therapy, for example. This seemed like an opportunity to me to use my skills in molecular biology to help understand these lymphomas better. So, that was the basic goal.
PEGGY: Lou's big idea was to look at the molecular characteristics of cancer, which might lead to a better understanding of the disease, which might lead to better things for the patient. And then he had this other, even bigger, idea.
[SONG: "Beach"]
LOU: I sort of had the sense that I didn't want to devote my entire life to one gene that seemed a little too small. What I wanted to do was to look at all the genes.
PEGGY: Lou wanted to study all the genes in order to understand cancer. If you're familiar with the field of cancer genomics now, this might not seem like a big idea. Because this is pretty much what researchers do routinely today: for all the genes, they look for mutations, track gene expression, and lots more--using a technology called next-generation sequencing. But the only problem was, this was back in the mid 90s.
LOU: The genome sequence was close to being finished at that time. We didn't know where all the genes were.
PEGGY: Not only was there no next-gen sequencing technology, there was no reference human genome.
LOU: we take the genome for granted, you know the genome is 3 billion base pairs. At the time, you know, that seemed daunting. And most people certainly had not been taught anything about the genome in medical school.
PEGGY: So 20 plus years ago, the idea of looking at the genomes of cancer seemed impossible. It seemed out there. In fact, when Lou took this idea to his colleagues, they said things like…
LOU: "Why would you want to do that?" You know? It was just very foreign to the standard molecular biologist to start thinking about the whole genome.
PEGGY: So Lou was met with some skepticism from the community. But he continued to think about ways he could study the genomes of cancers. He didn't get very far until by chance, a friend came back from a trip to Stanford and said…
LOU: "Well you'll never believe what Pat is doing. He is able to look at the messenger RNAs of thousands of genes at the same time by printing the DNA for those genes in like, tiny, tiny little microscopic dots on a glass microscope slide.” So, you could squeeze, you know, 10,000 spots on one microscope slide. And this lightbulb went off and I said, "That's exactly what I want to do."
PEGGY: That Pat was Pat Brown, the inventor of the microarray--an important technology for genomics that predates next-gen sequencing. With the microarray, you went from measuring expression levels for just a few genes to tens of thousands of genes at one time. Lou had found a key starting piece for his project. But he still had a massive amount of work ahead and needed a partner. He recalls interviewing students for his project.
LOU: I thought it was a great project, so I gave my best sale job for this. And one after the other, the students turned me down and went to other labs.
PEGGY: But finally, Lou was able to find someone who shared his large-scale way of thinking.
LOU: Ash Alizadeh, who happened to be from Stanford. And he said, "Cool. I'd love to do that."
PEGGY: With Ash on board, they got started. Their goal was to measure the expression levels of all the genes for diffuse large B cell lymphomas, a type of blood cancer. But how do you do something for all the genes, when you don't even know what all the genes are? They were going to have to design and build a microarray from scratch, and this was going to be a huge endeavor.
[SONG: "Traveling in Your Mind"]
PEGGY: First they needed to figure out what genes exist. Imagine trying to design a phone survey for 18,000 people, but first you have to write the phone book.
LOU: So, what we did was take a lymphoma cell and we isolated the messenger RNAs.
PEGGY: All this Lou is explaining is basically a lot of lab work.
LOU: Make a library, thousands of plasmids.
PEGGY: Time-consuming manual labor that had to be done for each sequence.
LOU: Old-school. No Illumina sequencer back then.
PEGGY: Followed by careful detective work and painstaking organization to figure out the location and name of each sequence, if possible.
ASH ALIZADEH: I'm not sure I want to totally admit, I was spending a ridiculous amount of time in the lab. It was a lot of work.
PEGGY: This is Ash.
ASH: I'm Ash Alizadeh, I'm a medical oncologist and cancer geneticist at Stanford University. I'm an associate professor in the Department of Medicine here. I take care of patients with lymphomas clinically and I do cancer genomics work in the research lab.
PEGGY: Ash spent a few years at the NIH through a fellowship--a few years mostly spent inside the lab.
ASH: I think I may have seen Smithsonian and the White House.
PEGGY: Ash spent long hours getting the sequences of genes expressed in a B cell, and trying to figure out what they were. Going back to the phone book analogy, it was like only having the phone numbers but not the person they belonged to. Again, with no reference human genome, this was a huge endeavor.
LOU: We had all these sequences, but then we didn't know what they were. And that's when we started blasting them against the UniGene database.
[SONG: "Traveling in Your Mind"]
PEGGY: Lou is describing taking each one of their sequences, and looking for a match within a very new gene database at the NCBI, the National Center for Biotechnology Information.
LOU: We started one by one, doing it manually, and this was very cumbersome. So, Ash was pretty enabled. He was a computer geek in high school. And so, he actually wrote a small computer script…
PEGGY: To automatically run the comparison.
LOU: So then one day I get this email…
PEGGY: from the Director of NCBI…
LOU: and he said, "Some computer mapped to your laboratory is pinging our computer servers repeatedly and taking them down. You must cease and desist."
ASH: Yes I did a lot did a lot of uh, yes I may have done that.
PEGGY: Ash actually recalls two different incidents of cease and desist emails, each taking down different sets of NCBI machines.
ASH: So if you can get something one at a time by going to a web browser of course I can write a three line piece of code that pulls down the whole thing. I started a laptop in my dorm room and another one at my bench at the lab and another one in another office and I came back to my desk at Stanford the next morning to have a couple of very important people at my desk because the entire Stanford campus had been unplugged from the NIH.
PEGGY: So Ash crashed NCBI resources and caused quite a stir. Twice.
ASH: It was quite embarrassing of course.
PEGGY: But after each time, Ash was able to apologize and explain their work. And make a plan that didn't take down servers. And finally, after about two years, they had sequenced and labeled thousands of sequences for…
LOU: what we fondly termed, the Lymphochip.
[SONG: "Beach"]
LOU: Took one to two years to fully annotate all those 18,000 spots on the microarray. So, it was a bit of a labor of love to do that.
PEGGY: Very repetitive.
LOU: It was very repetitive, but interesting. It was repetitive but like eating peanuts because you'd blast this gene and you say, "Oh. That's that gene. I didn't know we had that one on the microarray. Cool."
PEGGY: Another part of the labor of love was physically printing their Lymphochip. It took a lot of work in the lab to prepare all those sequences from their B-cell "phone book" for printing, involving…
ASH: Picking bacterial colonies for tens of thousands of cDNAs by hand, and then mini prepping and sequencing and PCRing, and then building robots.
[SONG: "Traveling in Your Mind"]
PEGGY: Building robots: putting together machinery to print thousands of tiny spots of DNA on a glass slide less than 1 by 3 inches. This was something they learned to do through their collaboration with Pat Brown.
LOU: Ultimately it's a tedious process. It was very, sort of a Rube Goldberg kind of machine that people had put together. The tip would jam or the tip would clog up with a little piece of schmutz so you had to stop the robot and take the tip off. We had a hundred microarrays in a batch that we made by hand and you decided one by one how you were going to use those. If you used them up you had to go back and make a hundred more and that was painful.
PEGGY: Designing and printing the physical glass slide was one aspect of building the Lymphochip, but what about the data? With 18,000 spots on a slide, how do you keep track of what spot is what gene?
LOU: That was the beginning of where I sort of developed databasing skills. So, without a database, you were nowhere with all this data. And there was a tool I learned how to use, actually with my kids, called HyperCard. Tthey made games, you could chase a ball around the thing. And it occurred to me that I could make one page for every spot on the microarray. So, that's how I made my first database, was using this kids program, HyperCard.
PEGGY: They actually moved on from using a kids game to store their genomic information after they established another collaboration…
LOU: With the Center for Information Technology, CIT, at the NIH and a guy named John Powell and John was a real computer scientist.
PEGGY: And with the database to hold their information, they still had to figure out how to analyze the data.
LOU: This was the first time that people started having a lot of data, a lot of genomic data. You had thousands of data points from one experiment.
[SONG: "Traveling in your Mind"]
ASH: We really didn't know how to analyze the data right, each chip has thousands of features, and these features now are measured as red and green and they turn out as a set of numbers that you can open up in Excel, but then you do the next chip and you have another bunch of numbers in Excel, and how do we analyze this high-dimensional data?
LOU: I think we were measuring about 10,000 genes.
PEGGY: 12,692, to be exact.
LOU: Which ones tell you something interesting, informative about the nature of that tumor?
ASH: A really talented person by the name of Mike Eisen, I ended up working very closely with Mike and learning from him. He built some amazing tools for looking at the data.
PEGGY: And then, they came upon a part of their work that is still a big hurdle in cancer studies today.
ASH: I had some pretty tense conversations with Lou that hey we spent a lot of time for this we gotta now be able to get the human tumors.
PEGGY: If you want to measure gene expression on human tumors, you're going to have to find some human tumors from real patients. So far, all their work had been using cell line models that are easy to get, but not actual tumors direct from patients.
ASH: Now we had this amazing tool in our hands that we had just built, we needed human specimens and it was tough to collect those specimens.
LOU: Through the help of the Rick Klausner at the National Cancer Institute, he put me in touch with some groups especially at the University of Nebraska that specialized in treating patients with lymphoma. And then we also had some tumor samples from patients at Stanford.
PEGGY: They were able to pull together 40 lymphoma sample thanks to Lou's skill of networking. These were precious samples that came from real people who had had this cancer.
ASH: I remember how anxious I was when I would get these tumors they would come in the mail on dry ice and I would think wow this human being had this tumor taken five ten years ago and they're alive or they're dead and now I have this piece of their tissue in my hands and I have to make sure I don't screw this up.
[SONG: "Beach"]
ASH: That was pretty stressful. Nobody had done that on these chips before to look at human tissues at that resolution so it was really exciting.
PEGGY: Things were getting very serious, and it was exciting. After all this work designing and building this new tool, finding the right collaborators, developing the data system and analytical methods, acquiring real patient samples, Lou and Ash were finally ready to get to the part that was the whole point of their work. To use…
LOU: Molecular biology to help understand these lymphomas better.
PEGGY: And the results?
ASH: Something very surprising happened when we were comparing the samples to each other. Which was a group of the samples from these patients clustered very closely with these tonsillar b-cells that I had purified from tonsils from children who had their tonsils removed.
PEGGY: One of the places you can get "normal" B-cells is from tonsils, and they tested some as a comparison for their cancerous B cells. And some of their tumors looked a Lot like these tonsils, which was surprising.
ASH: So surprising that we had to repeat it so many times to convince ourselves that it was meaningful. We repeated a couple times and showed it to Lou and Lou immediately knew what it meant.
PEGGY: To understand Lou's interpretation, we need to step back and go over a little blood biology.
[SONG: "Traveling in Your Mind"]
LOU: B-cells are in the blood, and then when they encounter a foreign antigen, they start dividing, become activated, and then ultimately form a new microenvironment in your lymph nodes called the germinal center. And it was becoming clear from other work that I was doing in my laboratory and from other laboratories that some of the human lymphomas seem to be derived from these germinal center B-cells as opposed to the blood B-cells.
PEGGY: There are different subtypes of B cells spanning different stages of development. The idea was that some cancers, even though they all looked the same under the microscope, came from these different B-cell subtypes. So they focused on the genes that distinguished a blood B cell from a more differentiated, germinal center B cell.
LOU: And with that set of genes, we found two groups of the lymphomas. And so, that was the first indication that there might be subtypes of diffuse lymphoma.
PEGGY: And did that cluster along response to treatment?
LOU: It did. So, that was really interesting.
PEGGY: They observed two patterns among their 40 samples. And which pattern a tumor had seemed to matter a lot for survival.
LOU: Nowadays you would want to have hundreds of patients to do this kind of work, but that’s what we had. And so, we took the survival data and looked at it for these two groups, and it was strikingly different.
PEGGY: Lou and Ash named the lymphoma subtype that looked like activated B cells “ABCs”, and the ones that looked like germinal center B cells “GCBs”.
ASH: Some look much more like the germinal center stage in the development of the B cell than others and that was a kind of a strange concept at the time.
PEGGY: Ash repeated a lot of experiments to collect more data and also help convince himself that what they were seeing was not a fluke. One of these experiments was checking the gene expression of more of these "normal" tonsil B cells.
ASH: I don't think Lou really knows this story so you're hearing it for the first time
[SONG: "Traveling in Your Mind"]
ASH: They're pretty rare cells. They're pretty fragile cells and to purify them takes a lot of time, so I spent a lot of time in the cold room sorting them out of pediatric tonsils and pooling them from a bunch of surgeries and to get enough numbers. And when I was ready to load basically the cDNA from the probe that I had made from several months of my life to put on the microarray, the tip of my pipettor fell off the pipette.
PEGGY: [Gasp]
ASH: And it fell on a bench where another graduate student had spilled a lot of popcorn and I just, my heart just broke that this my experiment is just ruined because of the popcorn that Paul has eaten and I dropped all my probe here and I said screw it I'm going to pick this probe off this yucky bench and put it on this glass slide and that glass slide is in the Nature paper.
PEGGY: The data from that slide is in Figure 4 of the paper, by the way.
ASH: It did confirm the original result, everything worked out.
PEGGY: [Laughing] I don't know, Ash, I think I need to notify Nature that…
ASH: [Laughing] Popcorn may be involved, yes, yes.
PEGGY: They also spent time checking their statistics and analyses, and all of this careful work cost them time. When Lou and Ash were wrapping up their work…
ASH: Um, you know we were a little bit scooped.
PEGGY: Getting scooped—having someone else publish similar work first—can affect how your work is seen and published. It’s a common source of stress for researchers. So a few other groups had been working on projects similar to Lymphochip, and one group from MIT published a microarray study on leukemia. One difference, the group used…
ASH: Commercial microarrays that were synthesized.
PEGGY: The commercial chips from a company had much fewer genes.
ASH: They were asking different questions but using similar tools and were around the same ideas.
PEGGY: I asked Lou about this, and he said a major difference is the other group was describing features of already known subtypes. Not discovering new ones, as Lou and Ash were doing. But still, to be scooped, or kind of scooped, right at the very end. This can be a researcher's worst nightmare.
ASH: That's often the way science works but I was a student I remember being very stressed out about it.
[SONG: "Beach"]
ASH: It was good to see that those other projects were able to use the same ideas and move forward.
PEGGY: So taking a semi-maybe-scoop with grace, knowing there were others doing similar work, Ash and Lou were at a point where they had to decide if they were finally ready to submit their work to a journal, and what exactly they were going to state--had they gathered enough evidence to claim they had discovered subtypes of their lymphoma--their ABC and GCB subtypes?
ASH: I don't think I had the scientific maturity to know that this was a risk worth taking and I think Lou of course did and to say that we've discovered cancer subtypes using this method was kind of a big statement to make.
LOU: Though we had a small number of patients, I had to make a decision when we went to publish this, you know, I could be wrong about this. I could be dead wrong because, you know, we could study the next 100 patients, and they would have a completely different survival. But, again, no guts, no glory. So, I decided to put that very preliminary, early data in our paper. It was solid data, but it just hadn’t been confirmed, and, of course, I said, “This needs to be confirmed in other studies.” I said that in the paper, but, nonetheless, I was definitely going out on a limb.
PEGGY: So Lou and Ash went out on a limb, submitted their study with only 40 patients to the journal Nature and waited for reviews. The fate of their beloved Lymphochip now rested with reviewers. Reviewers who, maybe they wouldn't understand the Lymphochip. Or maybe couldn't get on board with a genomic way of thinking. Or maybe they were on board but didn't feel the work was novel because of the scoop.
PEGGY: Was there pushback from the reviewers?
LOU: They were universally enthusiastic and this thing sailed right in and the editors of Nature loved it.
[SONG: "Ending"]
LOU: You know, the whole review process took like two months, it was really, really fast. I wish all my reviews were like that.
PEGGY: After an unsually lengthy time doing the research, and an unusually short time in review, the Lymphochip was published in Nature in February 2000.
LOU: And that article has been cited the most of anything that I’ve ever written, that’s I think up to about 6,000 citations.
PEGGY: Over 9,000 times, as of July 2019, according to google scholar. It turned out to be a seminal paper in cancer genomics. Getting the paper published is a major milestone for the researcher, but it is not the ultimate goal in science. Would Lou and Ash's biological discovery about ABC versus GCB lymphomas hold up?
LOU: Now, it turns out I was absolutely right. And through the years, every time people have looked at the ABC lymphomas, they are the difficult ones to cure with chemotherapy. And when you identify the patients with the GCB type, they do much better.
PEGGY: How was Ash able to stay motivated over the entire course of this project, working non-stop for about four years in total?
ASH: What Lou put at the end of this is: can you imagine this, can you see this, if you build it what you can do with it? The idea of trying to build a tool that will take a huge amount of organization and infrastructure to put together but with the idea of this being a microscope to really look at what you're hoping to look at, it did give me the sense of the value of discovery driven research as a core part of the scientific process.
PEGGY: Ash believed in this vision of a genomic tool, and the experience helped shape him as a scientist. And for Lou, who had done years of work already…
LOU: It was a turning point for me and many other people. And so, I starting thinking of all sorts of ways that you could use DNA microarrays. My thinking changed dramatically as a result of this first paper, because everything that I cared about doing was high-throughput from that point.
PEGGY: You might remember, in addition to doing lab research, Ash also takes care of patients.
ASH: Getting out of a patient's room who had just seen with the same disease that I you know was studying with Lou 20 some odd years ago and realizing that that those findings at the time still haven't changed the lives of our patients directly today.
[SONG: "Beach"]
ASH: Even though we made a major biological discovery, it's really much harder to change the way we take care of the disease. It’s a complicated issue and this is something that I worked on for a lot of my life, so I feel pretty strongly about.
PEGGY: Lou and Ash made a major biological discovery and published in a big journal, but for patients with B cell lymphomas, the impact has actually been modest. Changing things for the patient remains a challenge.
LOU: This has been a journey for me because it is hard to change the practice of medicine. And even if you have something that looks great to you and is very informative scientifically, and even if it predicts the outcome of patients with therapy, it, you know, takes a while for the healthcare system to actually adopt it.
ASH: My lab, by and large, is a translational lab that's been focused on answering cancer biology questions with a very translational focus. Even after we start therapy for a patient with say an aggressive lymphoma it's very challenging to make an accurate assessment of how they're responding until months after we've started. The major focus has been on building tools and we stopped to show that these tools are useful for our patients and that's an entirely different challenge.
PEGGY: So Lou made a big discovery, changed the course of his career, and laid some important groundwork for the cancer genomics field.
[SONG: "Hope and Love"]
PEGGY: What did it take to do all this? Of course, a big, out there, idea.
LOU: What I wanted to do was to look at all the genes…
ASH: Before the number of genes in the genome were solved…
LOU: So really that thought was the beginning of the kind of thought that turned into precision medicine…
PEGGY: And two very dedicated, arguably obsessive researchers, years of hard work to transform that idea into something real.
ASH: Years of spending pretty much most of my life in the lab…
PEGGY: It took connecting with the right people.
LOU: Collaboration with Pat Brown…
ASH: The group of John Powell…
LOU: Rick Klausner…
ASH: Mike Eisen…
PEGGY: All very big names in the field today, and there were of course even more people involved. It took creativity and resourcefulness, in order to figure out how to do some things for the very first time.
LOU: So, that's how I made my first database, was using this kids program, HyperCard.
ASH: I can write a three line piece of code that pulls down the whole thing.
PEGGY: Careful checking of work.
ASH: We had to repeat it so many times to convince ourselves that it was meaningful.
PEGGY: Bravery.
LOU: No guts, no glory, so I decided to put that very preliminary early data in our paper.
PEGGY: And a true passion for the work.
LOU: We had 100 microarrays in a batch that we made by hand…
ASH: I spent so much time in building these tools…
LOU: Again, a labor of love…
PEGGY: And what about the personal relationship between Lou and Ash?
LOU: I'm very proud of the fact that ash has now grown up and he is now a professor at Stanford. He's one of my very successful mentees.
ASH: Oh, it's a great honor for him to say something like that. I of course owe a huge debt of gratitude to all the energy and effort he invested in my career development and in mentoring me.
PEGGY: Ash says of the time spent in Lou's lab…
ASH: I remember them as some of the best years of my training.
PEGGY: It’s been more than 20 years since Lou started building his Lymphochip and cancer genomics research took off. The Lymphochip has since been retired and the field has moved on to different technologies. But the tools and infrastructure that genomics researchers have today, they are here in part because of the foundational work done by Lou, Ash, and others like them. Here's Lou on where all this has lead us:
LOU: At the current time we're doing more clinical trials we're doing combinations of drugs to improve the activity, but that the drugs we're combining are not chemotherapy drugs, they're targeted therapy drugs. So that's really where precision medicine takes you.
PEGGY: And how does that feel to have 20 years of work building up towards…
LOU: It feels great. It feels it feels like, one thing you learn in medical school is your job is to make human disease either better or go away. You develop this real desire to help people and what I’m hoping is that we actually, through some of our studies, will have helped some patients with cancer.
PEGGY: Big thanks to Lou Staudt and Ash Alizadeh for sharing their personal stories about working on a big idea that some people thought were crazy. And taking the time to be part of this podcast. For more information about this episode, including some fun pictures of Ash, visit our website at cancer.gov/personalgenomics.
[SONG: "Friend to Friend"]
PEGGY: If you have questions about cancer, comments about this podcast or would like to share your own research story, email us at NCIinfo@nih.gov. Be sure to mention the Personal Genomics podcast. If you like the show and want to hear more, please subscribe, share it with a friend, and leave us a review on iTunes. Personal Genomics is produced by me, Peggy Wang, with help from CCG staff. We are a production of the US Department of Health and Human Services, National Institutes of Health, National Cancer Institute. By the way, speaking of getting met with skepticism for your ideas, when I talked to Lou about this podcast...
LOU: You're going to have to edit this because people are going to be bored.
PEGGY: I'm Peggy Wang, see you on the next episode of Personal Genomics.