Computational Human High-grade Glioblastoma Multiform (GBM) Interactome - miRNA (Post-transcriptional) Layer
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Reference
Sumazin et al. (Cell, 2011)
Data
- Raw/Analyzed Data (.zip file)
The Human High-Grade Glioma Interactome (HGi) contains a genome-wide complement of molecular interactions that are Glioblastoma Multiforme (GBM)-specific. HGi v3 contains the post-transcriptional layer of the HGi, which includes the miRNA-target (RNA-RNA) layer of the interactome.
Experimental Approaches
microRNA target predictions were obtained using a two-step machine learning approach. First, sites predicted using miRanda, PITA and TargetScan were scored by classifying sites against a gold standard of validated interactions using a Support Vector Machine (SVM). The SVM is trained on features including the normalized score from the predicting algorithm, conservation across mammalian genomes, and site location relative to the start and end positions of the 3’ UTR. Then co-expression, site scores, and modular site grammar were used to predict interactions with SVM. Features and parameters were selected using cross validation and produced high confidence predictions after retraining the SVM on the complete dataset.
Direct Reversal of Glucocorticoid Resistance by AKT Inhibition in Acute Lymphoblastic Leukemia (T-ALL)
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Reference
Piovan, Yu et al. (Cancer Cell, 2013)
Data
- Raw/Analyzed Data (GEO)
- Analyzed Data (.zip file)
The goal of this project is to identify key druggable regulators of glucocorticoid resistance in T-ALL. To this end, a reverse-engineered T-ALL context-specific regulatory interaction network was created from a phenotypically diverse T-ALL gene expression dataset, and then this network was interrogated using master regulator analysis to find drivers of glucocorticoid resistance. The T-ALL gene expression dataset represented many different biological conditions, genotypes, signaling and transcriptional states, thus providing significant variation in which to detect gene expression correlations.
The expression level of transcription factors is often a poor predictor of their activity and biological relevance. However, their activity at the protein level can be inferred by measuring changes in the gene expression of their targets between two phenotypes, for example between tumor and normal tissue. This approach, called master regulator analysis, has been used successfully to identify functional drivers of cancer in a number of studies. In this study, master regulator analysis was used to identify regulatory genes whose network targets were enriched in the signal transduction cascade (as reflected in a differential gene expression signature) associated with glucocorticoid resistance.
Microarray gene expression data used in network generation and master regulator analysis is available in Gene Expression Omnibus under accession number GSE32215.
Experimental Approaches
Reverse-Engineering of T-ALL Transcriptional Network (ARACNe)
For each gene in a list of regulatory genes (hubs), the ARACNe algorithm1,2 is used to measure the mutual information between that gene and all remaining genes in the dataset. First, a preprocessing run is performed in which a curve relating mutual information to significance is generated. Next, ARACNe is run using the adaptive partitioning algorithm, repeated 100 times with bootstrapping3. A key step after each run of ARACNe is the application of the Data Processing Inequality to remove indirect interactions, typically with a zero threshold. A final consensus network is reconstructed from the bootstrapped networks based on the support of each edge, using a null distribution obtained via permutations.
Gene expression data from 223 T-ALLs (Human U133 Plus2.0 Affymetrix microarray platform) was subjected to GC Robust Multi-Aarray normalization and non-specific filtering (removing probes with no Entrez id, Affymetrix control probes, and non-informative probes by IQR variance filtering with a cutoff of 0.5). A set of hub genes was defined including genes with annotated functions in signaling transduction (GO:0007165) such as kinases, phosphatases, ubiquitin ligases, etc. to establish a signaling factor-centered interactome at the transcriptional level. ARACNe was used to identify targets of these hub genes (that is, genes with significant mutual information with the hub genes). It was run using the adaptive partitioning algorithm with a p-value threshold of 1e-7, DPI tolerance of 0, and 100 rounds of bootstrapping.
Master Regulator Analysis (MARINa)
For master regulator analysis, a group of 22 glucocorticoid resistant and 10 glucocorticoid sensitive T-ALLs was selected from the larger dataset used in network generation. Genes were ranked by their differential expression between these two conditions. The MARINa algorithm uses Gene Set Enrichment Analysis (GSEA)4 to test the differential enrichment of the regulons of hub genes (network first-degree neighbors) in the rank of genes differentially expressed between glucocorticoid sensitive and glucocorticoid resistant samples5. For GSEA method the ‘maxmean’ statistic6 was applied to score the enrichment of the gene set in the glucocorticoid resistant vs. glucocorticoid sensitive leukemias and sample permutation was used to build the null distribution for statistical significance.
References
- Basso K, et al. (2005). Reverse engineering of regulatory networks in human B cells. Nature Genet. 37(4):382-390 (PMID: 15778709)
- Margolin AA, et al. (2006). ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context. BMC Bioinformatics. 7(Suppl.1):S7 (PMID: 16723010)
- Margolin A, et al. (2006). Reverse Engineering Cellular Networks. Nature Protocols 1(2):663-72 (PMID: 17406294)
- Subramanian A, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 102(43):15545-50 (PMID: 16199517)
- Carro MS, et al. (2010). The transcriptional network for mesenchymal transformation of brain tumors. Nature. 463(7279):318-25 (PMID: 20032975)
- Efron B and Tibshirani R. (2007). On testing the significance of sets of genes. The Annals of Applied Statistics. 1, 107-129.
Expression Profile of Neuroendocrine Tumor Cell-line Perturbed with Small Molecules
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Reference
Alvarez et al. (Nat Genet, 2018)
Data
We have developed a new precision oncology framework for the systematic prioritization of drugs targeting mechanistic tumor dependencies in individual patients. As a component of this project, we used drug perturbation assays to scan a library of compounds against the H-STS neuroendocrine tumor cell line. We evaluated each compound’s ability to invert the concerted activity of master regulator proteins that mechanistically regulate tumor cell state.
Experimental Approaches
H-STS cells were perturbed with a library of 107 small-molecule compounds at their corresponding ED20 concentration and one-tenth of it. Cells were lysed at 6 h and 24 h after small-molecule compound perturbation and total RNA was isolated for RNA-Seq analysis. Libraries for RNA-seq were generated with the TruSeq protocol (Illumina) and sequenced in a Hi-Seq 2500 instrument (Illumina). Summarized expression data resulting from these analyses are available from the Gene Expression Omnibus database (GSE96760).
PLATE-seq for Genome-wide Regulatory Network Analysis of High-throughput Screens
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Reference
Bush et al. (Nat Commun, 2017)
Data
Pooled Library Amplification for Transcriptome Expression (PLATE-Seq) is a new, highly scalable and multiplexed RNA-Seq protocol for barcoding and pooling cDNA libraries to substantially reduce the cost and complexity of multi-sample analysis. Here we describe its application to small molecule perturbation experiments using BT20 breast cancer cells. PLATE-Seq is part of a larger analysis pipeline that uses reverse-engineered gene regulatory networks, greatly reducing the sample sizes required to infer regulatory protein activity.
Experimental Approaches
We use automated liquid-handling to introduce lysis buffer, capture polyadenylated mRNA with an oligo(dT)-grafted plate, and deliver well-specific, barcoded oligo(dT) primers to every sample in a multi-well plate. After reverse transcription, the cDNA in each well contains a specific barcode sequence on its 5’-end and a common adapter, such that all samples can be combined into a single pool for purification and concentration. We then use Klenow large fragment for pooled second-strand synthesis from adapter-linked random primers. Because this polymerase lacks strand-displacement and 5’-to-3’ exonuclease activities, each cDNA molecule produces at most, one second-strand synthesis product containing the sample barcode. Finally, the pooled library is enriched in a single PCR prior to sequencing. The resulting libraries represent the 3’-ends of mRNAs and are sequenced to a depth of 0.5-2 million raw reads per sample.
To characterize the performance of PLATE-Seq, we conducted a fully automated, 96-well screen to profile BT20 breast cancer cells following treatment with seven well-characterized small-molecule perturbagens (plus DMSO controls) and 12 replicates per condition.
Pharmacological Targeting of Mechanistic Dependencies in Neuroendocrine Tumors
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Data
- Raw/Analyzed Data (SRA/GEO)
- Raw/Analyzed Data (.zip file)
We have developed a new precision oncology framework for the systematic prioritization of drugs targeting mechanistic tumor dependencies in individual patients.
In the course of validating the approach, we reverse-engineered a gene regulatory network using gene expression profiles from a cohort of 212 gastroenteropancreatic neuroendocrine tumors (GEP-NETs), a rare malignancy originating in the pancreas and gastrointestinal tract.
Experimental Approaches
Expression profiles were obtained for the samples by RNA-Seq. Expression data were normalized by equi-variance transformation, based on the negative binomial distribution with the DESeq R-system package (Bioconductor). The regulatory network was reverse-engineered using the ARACNe algorithm1,2. ARACNe was run with 100 bootstrap iterations using a set of 1,813 annotated transcription factors. Parameters were set to 0 DPI (Data Processing Inequality) tolerance and MI (Mutual Information) P value threshold of 10−8. The gene expression profiles are available on GEO as GSE98894. The resulting ARACNe regulatory network is included in this submission.
References
- Basso K, et al. (2005). Reverse engineering of regulatory networks in human B cells. Nat Genet. 37(4):382-90. (PMID: 15778709)
- Margolin AA, et al. (2006). ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. Suppl 1:S7. (PMID: 16723010)
Core Regulatory Elements of High-risk Neuroblastoma
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Reference
Rajbhandari, Lopez et al. (Cancer Discov, 2018)
Data
- Analyzed Data (.zip file)
This project provides a framework to determine the downstream effectors of the genetic alterations sustaining neuroblastoma subtypes.
The results show the critical effect of disrupting a 10-protein module centered around a YAP/TAZ-independent TEAD4-MYCN positive-feedback loop in MYCNAmp neuroblastomas, nominating TEAD4 as a novel candidate for therapeutic intervention.
Experimental Approaches
The subtype-specific candidate master regulator (MR) proteins were inferred by independent analysis of the National Cancer Institute’s Therapeutically Applicable Research to Generate Effective Treatments (TARGET) and the European Neuroblastoma Research Consortium (NRC) datasets. Algorithm for the Reconstruction of Accurate Cellular Networks based on an Adaptive Partitioning strategy (ARACNe-AP) was used to assemble cohort specific interactomes from the gene-expression profiles of neuroblastoma samples from TARGET and NRC datasets. Candidate MR proteins for each of the high-risk subtypes were then prioritized based on the enrichment of their transcriptional target genes in the subtype-specific signature using the Virtual Inference of Protein activity by Enriched Regulon (VIPER) algorithm.
Proteome-wide Signaling-network Analysis in Lung Adenocarcinoma
Principal Investigator
Andrea Califano, Ph.D.
Contact
Prem Subramaniam
Reference
Bansal et al. (PLoS One, 2019)
Data
- Analyzed Data (.zip file)
Phospho- Algorithm for the Reconstruction of Accurate Cellular Networks (pARACNe) is a novel algorithm for the systematic inference of protein kinase pathways.
In this study, pARACNe was applied to analyze published mass spectrometry-based phosphotyrosine profile data from 250 lung adenocarcinoma (LUAD) samples. The resulting network includes 43 Tyrosine Kinases (TKs) and 415 inferred, LUAD-specific substrates. The predictions were validated at >60% accuracy by Stable Isotope Labeling with Amino acids in Cell culture (SILAC) assays, including “novel” substrates of the EGFR and c-MET TKs, which play a critical oncogenic role in lung cancer.
Experimental Approaches
The Califano lab developed a new algorithm, pARACNe, for inferring signaling networks from phosphoproteomics data. This method reports the abundance of phospho-proteins as measured by high-throughput mass spectroscopy (MS) based assay, to reveal how kinases interact with their substrates. Inferring transcriptional regulatory networks with ARACNe relies on the gene-expression data that are usually continuous and non-sparse. Data obtained from methods, such as liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) via spectral counting, are typically discrete and very sparse. To handle these discrete abundances, the mutual information computation approach was modified from a kernel density estimation-based method to a histogram-based Naïve-Bayes approach.
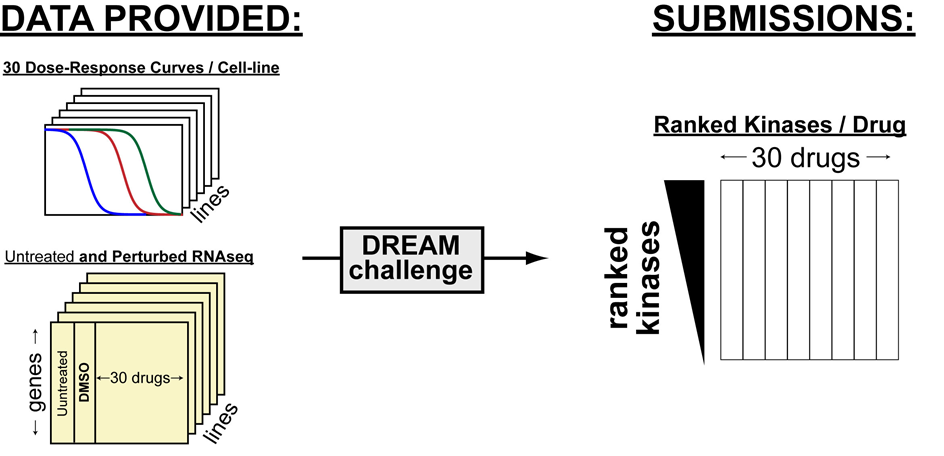
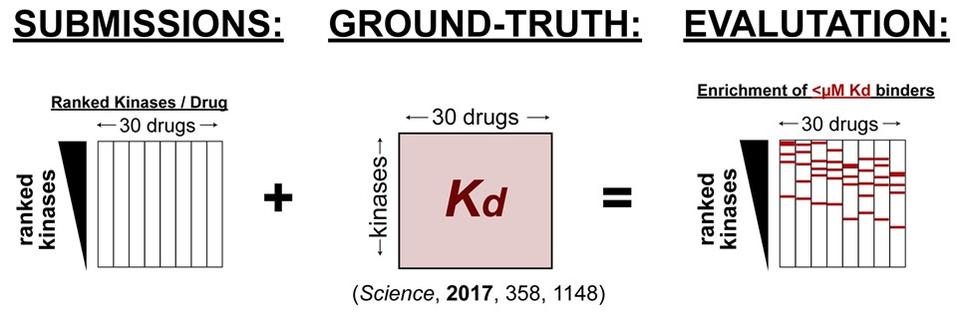
CTD² Pancancer Drug Activity Challenge
Principal Investigator
Andrea Califano, Ph.D.
Contact
Eugene Douglass
Reference
Douglass Jr. et al. (Cell Rep Med, 2022)
Data
- Analyzed Data (.zip file)
The goal of the CTD² Pancancer Drug Activity DREAM Challenge is to foster the development and benchmarking of algorithms to predict targets of chemotherapeutic compounds from post-treatment transcriptional data. The drug perturbational profiles on 11 cell lines and their dose-response curves for 32 chosen compounds with well-established targets will be provided to challenge participants, without revealing the identity of the drugs. These profiles will be removed from any public dataset and added back only after the challenge is completed. Transcriptional profiles for all the cell lines in which the compounds have been profiled have been provided to challenge participants, including the specific concentration at which the compound was titrated.
The package contains 2 metadata files, 22 data files, a README file that describes the data, and a COLUMNS file of descriptions of column headers shared by the 24 data files.
Experimental Approaches
Methods overview
This dataset was developed in collaboration between Columbia University Irving Medical Centers (CUIMC)’s High Throughput Screening Center (HTS), Sulzberger Genome Center and the Califano Laboratory in the Department of Systems Biology. Briefly, HTS handled cell-culture, cell-perturbation experiments and RNA extraction; the Genome Center performed RNA sequencing and the Califano laboratory performed data normalization, quality control, benchmarking and scientific and statistical analysis.
Compound titration curves
To determine the 48h ED20 of each drug, cell lines were plated into 96-well tissue culture plates, in 100 μL total volume, and incubated at 37°C. After 16 hours the plates were removed from the incubator and compounds were transferred into assay wells (1 μL) in triplicate. Plates were then returned to the incubator. After 48 hours the assay plates were removed from the incubator and allowed to cool to room temperature prior to the addition of 100 μL of CellTiter-Glo (Promega Inc.) per well. The plates were then mechanically shaken for 5 minutes prior to readout on the EnVision Multi-Label Reader (Perkin Elmer Inc.) using the enhanced luminescence module. Relative cell viability was computed using matched DMSO control wells as reference. ED20 was estimated by fitting a four-parameter sigmoid model to the titration results.
Perturbational profile generation
Using the previously described plating and perturbation procedure we perturbed each cell-line with each drug at its 48h ED20 value (measured above) or its CMax concentration. In order to optimize the clinical translation potential of the perturbation databases, we used the CMax, defined as the maximum plasma concentration after the administration of the drug at the maximum tolerated dose in patients, (whenever available from published pharmacokinetic studies), as an upper bound for the perturbation studies (Table S1). The mRNA from these cells was isolated and profiled by PLATESeq (Nat. Commun. 2017, 8, 105) at 24h after each perturbation.
Profile normalization
RNASeq reads were mapped for each well to the human reference genome assembly 38 using the STAR aligner,57 version 2.5.2b. Individual plates counts files were then combined, normalized and corrected for batch effects. First, individual counts files were combined across genes and ERCC2 spike-in counts removed, yielding the raw counts file for each cell-line experiment. Second, raw counts were quantile normalized and variance stabilized based on the negative binomial distribution with the DESeq R system package.59 To account for plate-based batch effects (which are common with drug-perturbed transcriptomic data) normalized expression was batch corrected using ComBat.60
OncoLoop: A Network-based Precision Cancer Medicine Framework
Principal Investigator
Andrea Califano, Ph.D.
Contact
Alessandro Vasciaveo
Reference
Vasciaveo et al. (Cancer Discov, 2023)
Data
- Raw/Analyzed Data (.zip file)
- Raw/Analyzed Data (SRA/GEO)
Prioritizing cancer treatment at the individual patient level remains challenging and performing co-clinical studies using patient-derived models in real-time is often unfeasible. To circumvent these challenges, we introduce OncoLoop, a precision medicine framework to predict drug sensitivity in both a human tumor and its highest-fidelity (cognate) model(s)—for contextual in vivo validation— by leveraging perturbational profiles of clinically-relevant oncology drugs. As proof-of-concept, we applied OncoLoop to prostate cancer using a series of genetically engineered mouse models (GEMMs) that capture the broad spectrum of disease states, including metastatic, castration-resistant, and neuroendocrine prostate cancer. Interrogation of published cohorts revealed that most patients were represented by at least one cognate GEMM-derived tumor (GEMM-DT), based on Master Regulator (MR) conservation analysis. Drugs recurrently predicted to invert MR protein activity in patients and their cognate GEMM-DTs were successfully validated, including in two cognate allografts and one patient derived xenograft (PDX). OncoLoop is highly generalizable and can be extended to other cancers and other pathologies.
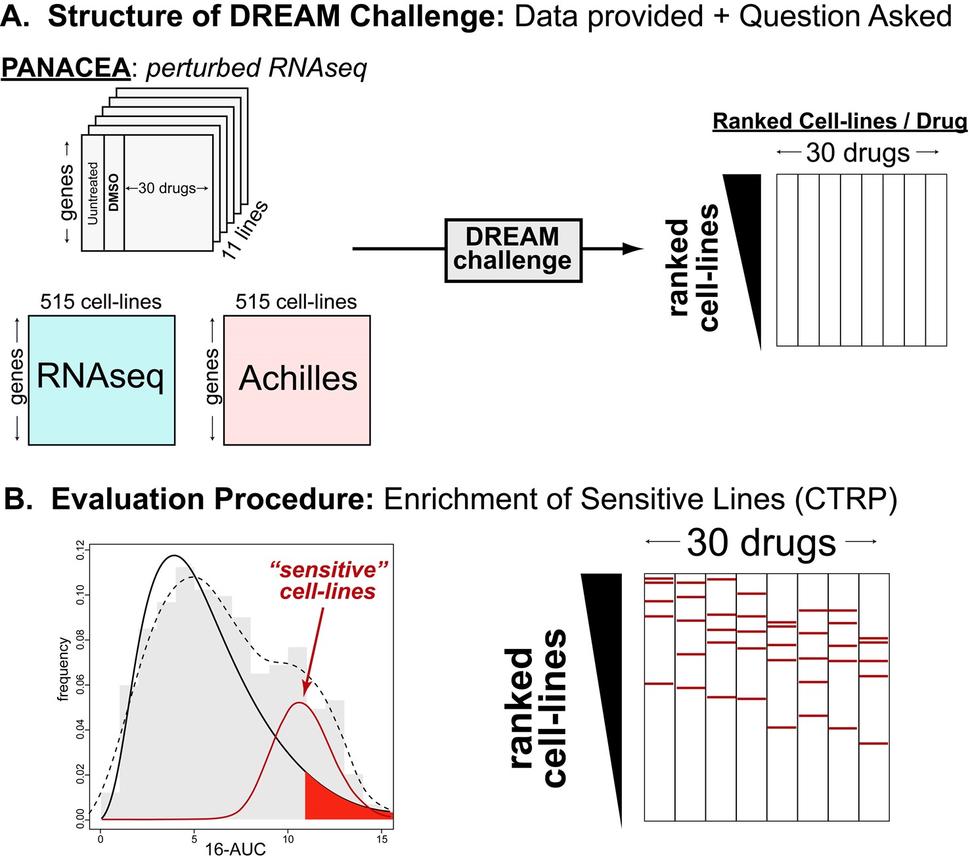
CTD² Pancancer Chemosensitivity Challenge
The goal of the CTD² Pancancer Chemosensitivity DREAM Challenge is to foster the development and benchmarking of algorithms to predict drug-sensitivity using post-treatment transcriptional data.
The drug perturbational profiles on 11 cell lines and for 30 chosen compounds will be provided to challenge participants, without revealing the identity of the drugs.
In addition, basal RNAseq and Achilles RNAi dependency data will be provided for 515 cell-lines which also occur within the CTRP drug-sensitivity data set.
Participants will be asked to use this data on drug-gene perturbations (PANACEA) and gene expression (RNAseq) and dependency (Achilles) to predict drug sensitivity for 30 drugs across 515 cell-lines.
Predictions will be evaluated by looking at the enrichment of “sensitive cell-lines” within the ranked predictions. “Sensitive cell-lines” are defined by fitting raw CTRP AUC data to a bimodal normal mixture model and establishing a threshold for sensitivity at a p-value of 0.5 with respect to the most resistant sub-population.
The package contains 4 metadata files, a README file that describes the data, and a COLUMNS file of descriptions of column headers shared by the 48 total data files.
Experimental Approaches
Methods overview
This dataset was developed in collaboration between Columbia University Irving Medical Centers (CUIMC)’s High Throughput Screening Center (HTS), Sulzberger Genome Center and the Califano Laboratory in the Department of Systems Biology. Briefly, HTS handled cell-culture, cell-perturbation experiments and RNA extraction; the Genome Center performed RNA sequencing and the Califano laboratory performed data normalization, quality control, benchmarking and scientific and statistical analysis.
Compound titration curves
To determine the 48h ED20 of each drug, cell lines were plated into 96-well tissue culture plates, in 100 μL total volume, and incubated at 37°C. After 16 hours the plates were removed from the incubator and compounds were transferred into assay wells (1 μL) in triplicate. Plates were then returned to the incubator. After 48 hours the assay plates were removed from the incubator and allowed to cool to room temperature prior to the addition of 100 μL of CellTiter-Glo (Promega Inc.) per well. The plates were then mechanically shaken for 5 minutes prior to readout on the EnVision Multi-Label Reader (Perkin Elmer Inc.) using the enhanced luminescence module. Relative cell viability was computed using matched DMSO control wells as reference. ED20 was estimated by fitting a four-parameter sigmoid model to the titration results.
Perturbational profile generation
Using the previously described plating and perturbation procedure we perturbed each cell-line with each drug at its 48h ED20 value (measured above) or its CMax concentration. In order to optimize the clinical translation potential of the perturbation databases, we used the CMax, defined as the maximum plasma concentration after the administration of the drug at the maximum tolerated dose in patients, (whenever available from published pharmacokinetic studies), as an upper bound for the perturbation studies (Table S1). The mRNA from these cells was isolated and profiled by PLATESeq (Nat. Commun. 2017, 8, 105) at 24h after each perturbation.
Profile normalization
RNASeq reads were mapped for each well to the human reference genome assembly 38 using the STAR aligner,57 version 2.5.2b. Individual plates counts files were then combined, normalized and corrected for batch effects. First, individual counts files were combined across genes and ERCC2 spike-in counts removed, yielding the raw counts file for each cell-line experiment. Second, raw counts were quantile normalized and variance stabilized based on the negative binomial distribution with the DESeq R system package.59 To account for plate-based batch effects (which are common with drug-perturbed transcriptomic data) normalized expression was batch corrected using ComBat.60
NaRnEA: An Information Theoretic Framework for Gene Set Analysis
Principal Investigator
Andrea Califano, Ph.D.
Contact
Zhongming (Lucas) Hu
Reference
Griffin et al. (Entropy (Basel), 2023)
Data
We created Nonparametric analytical Rank-based Enrichment Analysis (NaRnEA) to facilitate accurate and robust gene set analysis with an optimal null model derived using the information theoretic Principle of Maximum Entropy.
Experimental Approaches
All experimental methods necessary for reproducing the results of the manuscript may be found online in the manuscript (https://www.mdpi.com/1099-4300/25/3/542); all code may be found in the CCG GitHub repository.
Systematic Elucidation and Pharmacological Targeting of Tumor-Infiltrating Regulatory T Cell Master Regulators
Principal Investigator
Andrea Califano, Ph.D.
Contact
Luca Zanella
Reference
Obradovic et al. (Cancer Cell, 2023)
Data
- Raw/Analyzed Data (.zip file)
Due to their immunosuppressive role, tumor-infiltrating regulatory T cells (TI-Tregs) represent attractive immuno-oncology targets. Analysis of TI vs. peripheral Tregs (P-Tregs) from 36 patients, across four malignancies, identified 17 candidate master regulators (MRs) as mechanistic determinants of TI-Treg transcriptional state. Pooled CRISPR-Cas9 screening in vivo, using a chimeric hematopoietic stem cell transplant model, confirmed the essentiality of eight MRs in TI-Treg recruitment and/or retention without affecting other T cell subtypes, and targeting one of the most significant MRs (Trps1) by CRISPR KO significantly reduced ectopic tumor growth. Analysis of drugs capable of inverting TI-Treg MR activity identified low-dose gemcitabine as the top prediction. Indeed, gemcitabine treatment inhibited tumor growth in immunocompetent but not immunocompromised allografts, increased anti-PD-1 efficacy, and depleted MR-expressing TI-Tregs in vivo. This study provides key insight into Treg signaling, specifically in the context of cancer, and a generalizable strategy to systematically elucidate and target MR proteins in immunosuppressive subpopulations.
Experimental Approaches
See Methods Section of Published Manuscript at https://pubmed.ncbi.nlm.nih.gov/37116491/
A Transcriptome-Based Precision Oncology Platform for Patient–Therapy Alignment in a Diverse Set of Treatment-Resistant Malignancies
Principal Investigator
Andrea Califano, Ph.D.
Contact
Luca Zanella
Reference
Mundi et al. (Cancer Discov, 2023)
Data
- Raw/Analyzed Data (GEO)
- Raw/Analyzed Data (.zip file)
Complementary precision cancer medicine paradigms are needed to broaden the clinical benefit realized through genetic profiling and immunotherapy. We performed a first-of-kind evaluation of two transcriptome-based precision cancer medicine methodologies to predict tumor sensitivity to a comprehensive repertoire of clinically relevant oncology drugs, whose mechanism of action we experimentally assessed in cognate cell lines. We enrolled patients with histologically distinct, poor-prognosis malignancies who had progressed on multiple therapies, and developed low-passage, patient-derived xenograft models that were used to validate 35 patient-specific drug predictions. Both OncoTarget, which identifies high-affinity inhibitors of individual master regulator (MR) proteins, and OncoTreat, which identifies drugs that invert the transcriptional activity of hyperconnected MR modules, produced highly significant 30-day disease control rates. Predicted drugs significantly outperformed antineoplastic drugs selected as unpredicted controls, suggesting these methods may substantively complement existing precision cancer medicine approaches, as also illustrated by a case study.
Experimental Approaches
Generation of Gene Regulatory Networks: We have generated comprehensive molecular interaction networks (interactomes) using the Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNe) [1, 2], although other suitable algorithms may be used. The networks were reverse engineered by ARACNe from ≥ 100 RNASeq profiles of human cancer tissue from (a) The Cancer Genome Atlas (TCGA) and (b) for meningioma and neuroendocrine tumors, from Columbia University collected datasets. TCGA RNASeq level 3 data were downloaded from NCI Genomics Data Commons [3]. Raw counts were normalized and variance stabilized as implemented in the DESeq2 R-package [4].
VIPER Analysis: The Virtual Proteomics by Enriched Regulon analysis (VIPER) algorithm is a tool for the accurate inference of regulatory protein activity in tissue context-dependent manner [5-7]. VIPER leverages accurate tissue-specific gene regulatory networks, to measure differential protein activity from bulk or single-cell gene expression signatures (GES). For each cancer sample, we generate a differential gene expression signature (DGES)—the gene-wise relative expression to the distribution of the expression of that gene across 11,289 TCGA samples—and expressed as its quantile relative to the reference model. Next, VIPER computes enrichment scores for the targets of each regulatory protein in the DGES [5]. When cancer type specific networks are not available, we use an integrated network approach as implemented in metaVIPER [8, 9].
OncoTarget Analysis: Through the use of (a) DrugBank [10], (b) the SelleckChem database [11], (c) published literature, and (d) publicly available information on pharmaceutical company drug development pipelines, we have curated a refined list of 180 actionable proteins representing validated targets of high-affinity pharmacological inhibitors, either FDA approved or in clinical trials. This manually curated target-drug(s) database is dominated by signaling proteins and established oncoproteins, as expected. Pharmacological agents with narrow therapeutic indices—such as those targeting neurotransmitters, ion channels, and vasoactive drugs—were purposefully removed from the database as less likely to be successfully repurposed in oncology. OncoTarget simply analyzes the VIPER outputted protein activity measurements for these 180 actionable proteins, and provides a multiple-testing corrected significance value for the corresponding NES.
OncoTreat Analysis: For the current study, we broadly adapted OncoTreat to identify tumor checkpoint module (TCM)-inverter compounds. We identify sample-specific candidate MRs and the TCMs they comprise, by VIPER analysis of the sample’s DGES, compared to the set of TCGA samples (reference model). We assess drug effect by completing high throughput drug screens in relevant cognate cell lines with post-perturbation RNASeq using the multiplexed PLATESeq platform. Pharmacological agents were prioritized based on the statistical significance of the enrichment of the tumor sample’s TCM-activity signature (i.e., 25↑+25↓ MRs) in proteins inactivated and activated in drug vs. DMSO-treated cells.
OncoMatch, Cell Line and Patient-Derived Xenograft (PDX) Model Fidelity Analysis: Model fidelity was assessed based on the statistical significance of the TCM-activity conservation between a human-derived sample and a model-derived sample. The analysis was used to (a) select optimal cell lines for the generation of perturbational profiles that effectively track the activity of tested drugs on TCM proteins and (b) to assess the fidelity of PDXs prior to validation of drugs predicted from the human sample.
Establishment of PDX models and therapeutic drug testing: Fresh tumor tissue was fragmented and implanted subcutaneously into nonobese/severe combined immunodeficiency IL2Rg null, hypoxanthine phosphoribosyltransferase (HPRT)-null (NSGH) mice (Jackson Labs, IMSR catalog no. JAX:012480, RRID: IMSR_JAX:012480) and tumor engraftment monitored by visual and manual inspection.
Engrafted tumors were measured twice weekly with calipers and drug treatment initiated when tumor volume (TV) reached ~100 mm3 (TV = width2 X ½ length). Early passage animals (Passage 1 – 5) were used for all therapeutic studies.
Pharmacodynamic (PD) Assessments of TCM-inversion: Samples for PD assessment were procured from two mice per treatment arm. We performed RNASeq and subsequent VIPER on paired drug vs. Vehicle control-treated PDX tumor samples. TCM-inversion was assessed based on the statistical significance of the enrichment of the TCM-activity signature (i.e., 25↑+25↓ MRs of the patient tumor) in proteins inactivated and activated in drug vs. Vehicle control-treated PDX tumors, respectively.
References
1. Basso, K., et al., Reverse engineering of regulatory networks in human B cells. Nature genetics, 2005. 37: p. 382-90.
2. Margolin, A.A., et al., ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC bioinformatics, 2006. 7 Suppl 1: p. S7.
3. Zhang, Z., et al., Uniform genomic data analysis in the NCI Genomic Data Commons. Nat Commun, 2021. 12(1): p. 1226.
4. Love, M.I., W. Huber, and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550.
5. Alvarez, M.J., et al., Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet, 2016. 48(8): p. 838-47.
6. Bisikirska, B., et al., Elucidation and Pharmacological Targeting of Novel Molecular Drivers of Follicular Lymphoma Progression. Cancer Res, 2016. 76(3): p. 664-74.
7. Califano, A. and M.J. Alvarez, The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer, 2017. 17(2): p. 116-130.
8. Coutinho, D.F., et al., Validation of a non-oncogene encoded vulnerability to exportin 1 inhibition in pediatric renal tumors. Med (N Y), 2022. 3(11): p. 774-791 e7.
9. Ding, H., et al., Quantitative assessment of protein activity in orphan tissues and single cells using the metaVIPER algorithm. Nat Commun, 2018. 9(1): p. 1471.
10. Wishart, D.S., et al., DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res, 2008. 36(Database issue): p. D901-6.
11. FDA-approved & Passed Phase I Drug Library Contents. Available from: https://www.selleckchem.com/screening/fda-approved-passed-phase-i-drug-library.html.
Kinases Controlling Stability of the Oncogenic MYCN Protein
Principal Investigator
Andrea Califano, Ph.D.
Contact
Luca Zanella
Reference
Smith et al. (ACS Med Chem Lett, 2023)
Data
- Raw/Analyzed Data (.zip file)
MYCN is an oncogene that codes for a driver protein often found to be aberrantly activated or amplified in the cells of tumors with poor prognoses. We previously identified the natural products isopomiferin and pomiferin as powerful, indirect MYCN-ablating agents. In this work, we expand on their mechanism of action and find that casein kinase 2 (CK2), phosphoinositide 3-kinase (PI3K), checkpoint kinase 1 (CHK1) and serine/threonine protein kinase 38-like (STK38L), as well as STK38, work synchronously to create a field effect that maintains MYCN stability. By systematically inhibiting these kinases, we degraded MYCN and induced cell death. Additionally, we synthesized and tested several simpler and more cost-effective pomiferin analogues, which successfully emulated the compound’s MYCN ablating activity. Our work identified and characterized key kinases that can be targeted to interfere with the stability of the MYCN protein in NBL cells, demonstrating the efficacy of an indirect approach to targeting “undruggable” cancer drivers.
Experimental Approaches
The KINOMEscan conducted by Eurofins DiscoverX is a competitive binding assay. DNA-tagged kinases were incubated with biotinylated small molecule ligand docked to Streptavidin-coated magnetic beads and one of our submitted compounds (10 μM pomiferin, 15 μM compound 6, 15 μM compound 5, 5 μM CHIR124 and 10 μM AZD7762) in 1X binding buffer (20% SeaBlock, 0.17X PBS, 0.05% Tween 20 and 6mM DTT). Assays were conducted in 384-well plates in a final volume of 20 μL. Plates were incubated for one hour at room temperature while shaking, then washed with wash buffer (1X PBS, 0.05% Tween 20). The beads were resuspended in elution buffer (1X PBS, 0.05% Tween 20 and 0.5 μM non biotinylated affinity ligand) and incubated at RT while shaking for 30 minutes. The amount of kinase bound to the ligand in the eluates were measured via qPCR and compared to the DMSO control to determine %Ctrl.
% Ctrl calculation:
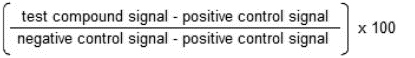
Subtype-selective Prenylated Isoflavonoids Disrupt Regulatory Drivers of MYCN-amplified Cancers
Principal Investigator
Andrea Califano, Ph.D.
Contact
Luca Zanella
Reference
Stokes et al. (Cell Chem Biology, 2024)
Data
Transcription factors have proven difficult to target with small molecules because they lack pockets necessary for potent binding. Disruption of protein expression can suppress targets and enable therapeutic intervention. To this end, we developed a drug discovery workflow that incorporates cell-line-selective screening and high-throughput expression profiling followed by regulatory network analysis to identify compounds that suppress regulatory drivers of disease. Applying this approach to neuroblastoma (NBL), we screened bioactive molecules in cell lines representing its MYC-dependent (MYCNA) and mesenchymal (MES) subtypes to identify selective compounds, followed by PLATESeq profiling of treated cells. This revealed compounds that disrupt a sub-network of MYCNA-specific regulatory proteins, resulting in MYCN degradation in vivo. The top hit was isopomiferin, a prenylated isoflavonoid that inhibited casein kinase 2 (CK2) in cells. Isopomiferin and its structural analogs inhibited MYC and MYCN in NBL and lung cancer cells, highlighting the general MYC-inhibiting potential of this unique scaffold.
Experimental Approaches
First Experiment (PLATESeq): To identify NBL MYCNA subtype-selective inhibitors, we systematically screened >5,000 compounds from three chemical libraries chosen for their enrichment of bioactive molecules, diversity in chemical structure, and for inclusion of compounds with known mechanisms of action. Compounds were initially tested at a single concentration and time-point (20 µM for 72 h) to eliminate those with no in vitro activity in NBL cell lines. Lethal compounds were rescreened across a five-point dilution series ranging from 20 µM to ~250 nM, to determine IC50 values in each of the four cell lines. By ranking compounds based on average IC50 values for each of the two subtypes, MYCNA or MES-selective compounds were identified. The top 90 compounds with the highest relative MYCNA-specific selectivity were then used to generate PLATESeq expression profiles following 24 hours of treatment in SK-N-Be2 cell lines at their respective IC20 concentration. Second Experiment (PLATESeq): top 15 compounds were screened with PLATESeq at 6 and 24 hours at multiple concentrations in three replicates Third Experiment (PLATESeq): In an effort to identify a potent analog of isopomiferin, we assembled a small collection of structurally-related analogs and known CK2 inhibitors, and tested them in SK-N-Be2 cells using PLATESeq TruSeq Experiment (RNASeq): To uncover kinase targets of isopomiferin and its functional analogs, we developed a novel algorithm, the Virtual-Inference of Kinase INhibiton by Gene regulatory networks (VIKING), to enable data-driven inference of drug targets based on activity dysregulation of protein-protein interactions (PPIs). VIKING was run using RNASeq (TruSeq) data on isopomiferin, pomiferin and its null analog, at multiple concentrations and timepoints.
Tumor-selective Effects of Active RAS Inhibition in Pancreatic Ductal Adenocarcinoma
Principal Investigator
Andrea Califano, Ph.D.
Contact
Luca Zanella
Reference
Wasko et al. (Nature, 2024)
Data
In this study 14 different RAF, MEK, or ERK inhibitors were tested in ASPC1 and PANC1 PDA cell lines. Panc1 and Aspc1 pancreatic cancer cells were cultured in white 96-well tissue culture-treated plates at optimized density, in 100 μl of Dulbecco's Modified Eagle Medium (DMEM) media supplemented with 10% fetal bovine. After 24 h of incubation, the plates were treated with following drugs: RAF inhibitors – Sorafenib, Dabrafenib, RAF709, PLX8394, GDC-0879; MEK inhibitors – Trametinib, Cobimetinib, Binimetinib, Selumetinib, Rafametinib; and ERK inhibitors – SCH772984, Ulixertinib, AZD0364, Ravoxertinib. Each drug was dosed at the concentration at which the cells were 80% viable after 48 h of treatment. After 24 h of treatment, the medium was replaced with 100 ml of FBS supplemented with 10% DMSO and the plates were frozen at −80 °C prior to PLATE-Seq. Detailed protocol for preparation of the automated PLATE-SEQ experiment was described by Bush et al. (Bush, Erin C., et al. "PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens." Nature communications 8.1 (2017): 105, https://www.nature.com/articles/s41467-017-00136-z). Samples were sequenced using PlateSeq approach.
Experimental Approaches
The PLATE-Seq FASTQ files were pseudoaligned to the GRCh38 human transcriptome and gene expression was quantified using kallisto (version 0.44.0), tximport package, and biomaRt package. The gene expression was quantified as both raw counts (i.e. sequencing fragments per genomic locus) and transcripts per million (i.e. sequencing fragments per genomic locus normalized for transcript/gene length and sample sequencing depth). Single sample differential gene expression signatures were computed independently for each one of the two cell lines and then integrated in order to derive a consensus MAPK signature. The z-score method was used to generate differential gene expression signatures of each drug-treated sample with respect to the DMSO-treated samples.
Raw data (FASTQs) and processed data (raw counts) for generation of experimental MAPK pathway gene expression signature have been deposited to GEO, accession number GSE252002.