Research
To transform the diagnosis and treatment of cancer using genomics, OCG implements research programs designed to illuminate cancer biology and generate rich community resources that benefit cancer researchers worldwide. Using a collaborative “team science” approach, OCG engages in three complementary sub-disciplines within cancer genomics: genome sequencing, functional genomics, and computational genomics.
-
Cancer Genome Sequencing
Cancer genome sequencing identifies alterations in cancer that contribute to cancer growth, metastasis, and recurrence. OCG's programs for characterizing cancers include The Cancer Genome Atlas and TARGET.
-
Functional Genomics Research
Functional genomics studies the role of particular cancer genes and pathways in the development and progression of cancer and harnesses that knowledge to develop therapeutic strategies against the disease. OCG's functional genomics research includes the Cancer Target Discovery and Development (CTD²) Network and the Human Cancer Models Initiative (HCMI).
-

Computational Genomics Research
Computational genomics uses statistical and computational approaches to derive insights from big data on cancer. OCG's work in this field helps develop tools for interpreting large, high-throughput datasets and make data secure and accessible for the research community.
-
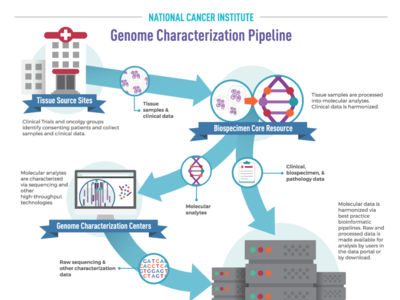
OCG's Genome Characterization Pipeline
NCI’s Genome Characterization Pipeline is an efficient workflow that transforms cancer tissue samples into rich, publicly available genomic and clinical data. By partnering with foremost experts in tissue processing, bioinformatics, and data science, OCG produces and shares high quality, harmonized raw and processed data with researchers worldwide.