CGCI Data Types and Availability
| Tumor Type/Data Use Limitation | Data Availability Status |
|---|---|
| Burkitt lymphoma/general research use (data use limitations for some cases are being confirmed) |
Pediatric whole genome sequencing, RNA-seq, and miRNA-seq data are available at NCI's Genomic Data Commons and Center for Cancer Genomics Data Coordinating Center. EBV sequences are available as BAM alignments from the public directory at the Data Coordinating Center. Pediatric study is complete. Adult study is in progress. |
| Non-Hodgkin lymphoma/cancer research only |
Open access (analyzed) and controlled access (upper-level files e.g., VCF or MAF) mRNA-seq data, whole exome, and whole genome sequencing data are available at the Center for Cancer Genomics Data Coordinating Center. Controlled access aligned sequence reads (BAM files) from mRNA-seq, whole exome, and whole genome sequencing data are available at the National Center for Biotechnology Information’s Sequence Read Archive. This project is complete. |
| HIV+ associated cervical cancer/general research use |
Controlled-access whole genome sequencing, RNA-seq, miRNA-seq, and targeted sequencing data are available at NCI's Genomic Data Commons. Controlled-access ChIP-Seq data and open-access methylation array data are available at the Data Coordinating Center. Both open and controlled-access higher level (analyzed) sequencing data are available at the Center for Cancer Genomics Data Coordinating Center. Note: The project includes cases from both HIV+ and HIV- cervical cancer patients. This project is complete. |
| HIV+ associated lung cancer/general research use and cancer research only |
To be determined. This project is in progress. |
| HIV+ associated diffuse large B cell lymphoma/general research use |
To be determined. This project is in progress. |
Cancer Genome Characterization Initiative (CGCI) researchers primarily use sequencing and, in some cases, other genome-based approaches to examine genomes and transcriptomes of tumors. With these in-depth analyses, they can detect cancer-associated alterations ranging from genomic rearrangements and copy number alterations in gene expression to single nucleotide insertion or deletion of small nucleotide mutations. By uncovering the full spectrum of alterations in tumors, CGCI researchers aim to elucidate which physiological pathways are disrupted in cancer. Finding the underlying molecular causes of cancer will, in some cases, inform better treatment strategies. Read below to learn about CGCI’s research approaches and the type of information they provide.
Central Pathology Review
Pathological diagnosis of tumors is important to confirm disease identification. To avoid any subjectivity associated with pathology reviews, and to allow efficient resolution of discrepancies, CGCI formed Pathology Review Committees consisting of three board‐certified pathologists. The committees ensure that the samples meet the requirements of CGCI projects. The tissue source sites submit either a Formalin-Fixed Paraffin-Embedded block or an appropriate number of unstained slides of tumor samples for pathology analysis. An H&E stained slide is used for initial tissue analysis for each submitted tumor to identify the location of the tumor. Immunohistochemistry stains are performed depending on the project and tumor subtype.
The pathology analysis can be done either individually or using tissue microarrays (TMAs). TMAs were used for the Burkitt Lymphoma Genome Sequencing Project.
Tissue Microarrays (TMAs)
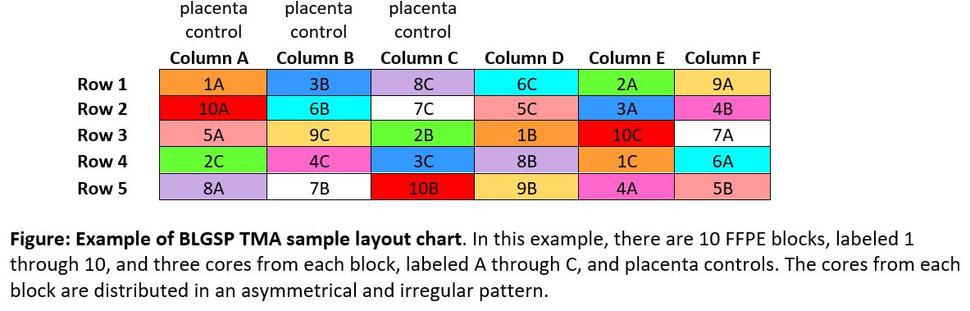
TMAs consist of paraffin blocks with separate tissue cores that are assembled in array fashion to allow multiplex histological analysis. TMAs for CGCI projects involve up to three tissue cores per case. In most TMAs, the tissue cores are placed according to a map (figure below) to remove possible pathology diagnosis bias. The coordinates indicating locations of specific cores on the slide are included in slide metadata. H&E stains from preliminary pathology analysis will be made available along with TMA images.
Immunohistochemistry and Fluorescence in situ Hybridization (FISH)
Antibody stains against the following proteins are used for respective projects as follows:
- Burkitt Lymphoma Genome Sequencing Project: CD3, CD10, CD20, BCL2, BCL6, Ki67, and MYC (FISH analysis)
- HIV+ Tumor Molecular Characterization Project
- Lung: p40 (optional in cases of suspected squamous cell carcinoma), TTF-1 (required for cases of suspected adenocarcinoma)
- Cervical: p16, p63 (optional for squamous cell carcinoma), p40 (optional for squamous cell carcinoma), BER-EP4 (optional for adenocarcinoma), MOC 31 (optional for adenocarcinoma), B72.3 (optional for adenocarcinoma)
- Diffuse large B-cell lymphoma: CD3, CD10, CD20, MUM1, BCL2, BCL6, Ki67, TP53, CD79a, EBER (RNA in situ hybridization analysis)
Burkitt Lymphoma Genome Sequencing Project Standard Operating Procedures Manual (PDF)
HIV+ Tumor Molecular Characterization Project Standard Operating Procedures Manual (PDF)