The Genomic Data Commons Turns 2: Progress in Clinical Tool Development
, by Louis M. Staudt, M.D., Ph.D.
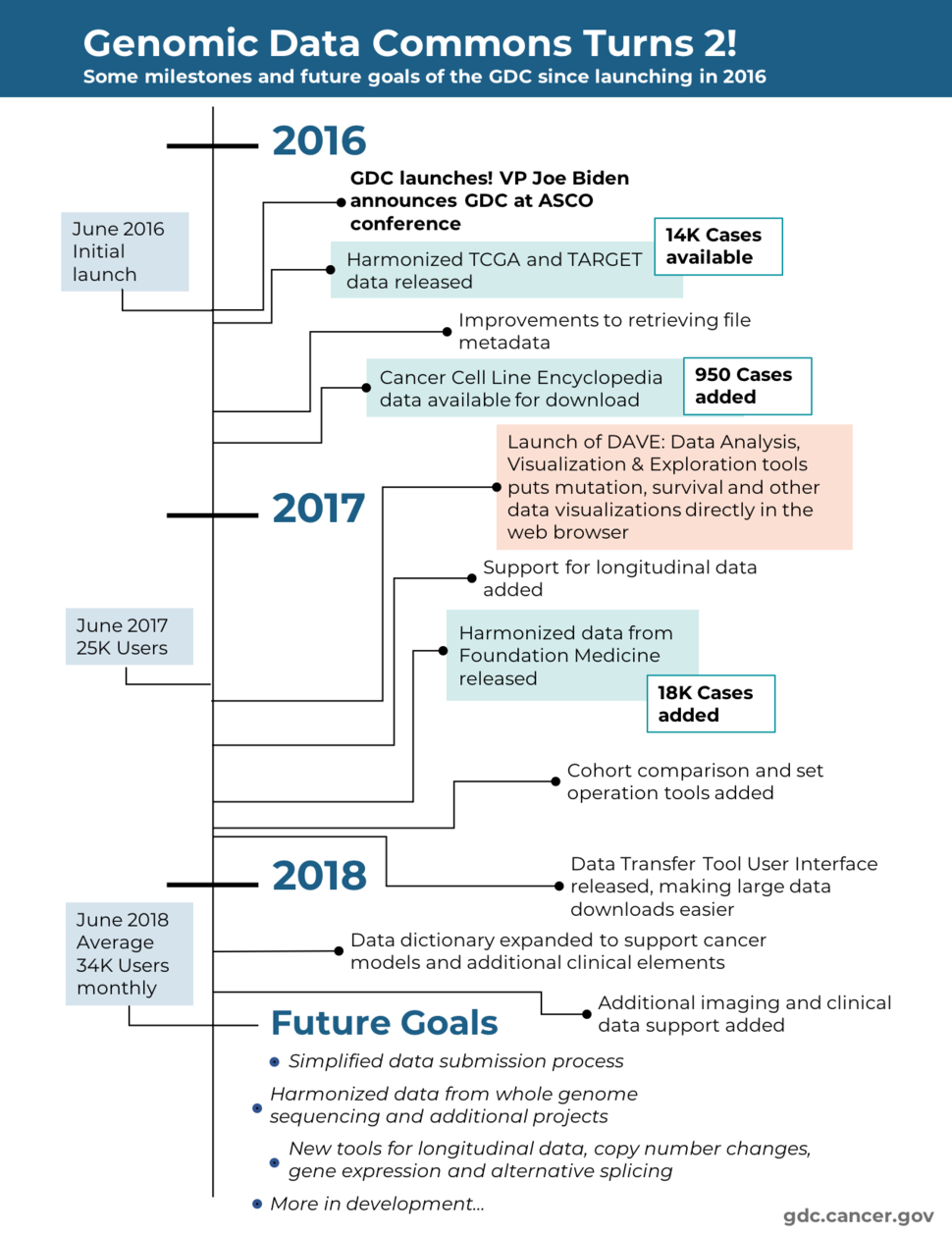
Two years ago, Vice President Joe Biden made a plea to the cancer research community for team science, collaboration, and greater sharing of data in order to advance precision medicine. With this goal in mind, he announced the launch of the Genomic Data Commons (GDC): our platform for bringing genomic and clinical data to anyone in the research community.
Since the launch, we’ve been working continuously to improve the GDC and build a system that is both valuable and accessible to all types of scientists and clinicians—by no means a trivial task. At the GDC’s second birthday, we’ve reached some important milestones and look to more challenges ahead in our quest to transform the research community.
Offering More Data in Raw and Analyzed Formats
We’ve collected and harmonized over 3 petabytes of genomic data so far. Now users can readily access a variety of analyzed data, including mutation calls, gene expression levels, copy number variations, and more. For the computationally inclined, raw sequencing data is available through controlled access. Over 32,500 cases of data from three major programs (TCGA, TARGET, and Foundation Medicine) are available and we are still adding more.
Building Interactive and Clinically Relevant Tools
While we originally launched as a data repository, our vision has been to create an interactive knowledge system that will readily connect the genomic characteristics of a particular patient’s cancer with clinical outcome.
A major step towards that vision was launching our Data Analysis, Visualization, and Exploration Tools, or DAVE, in June of 2017. With this resource, any person can open their web browser and instantaneously plot the frequencies and locations of mutations, survival curves, and other visualizations for the cases of their interest.
Since then, we’ve added tools for creating cohorts of patients and comparing vital status, gender, and other demographic characteristics. Users can also work with sets of genes or mutations. In our latest release, histology and pathology images can be viewed directly within the portal.
With less glamour but all of the importance, we’ve been making updates under the hood that improve the connection between genomic and clinical data and allow for clinically relevant analyses. These software and data model changes support the many features we are currently developing, including longitudinal analysis tools for tracking disease progression and therapeutic response at different timepoints for a patient.
Staying at the Forefront of Precision Medicine
While I am proud of the what we have built so far, a lot of work lies ahead. We are putting the finishing touches on tools for analyzing copy number, gene expression, and other types of data. We are streamlining the data submission process—a crucial part of building a platform meant to help researchers collaborate and share data. And we have a lot more in development.
In this rapidly developing field, we need your thoughts and ideas on how to make the GDC a valuable resource for you. We’ve heard a lot of great feedback so far from the research community, such as better integration between DAVE and downloading data (release notes for that update here) and bridging the GDC with drug compound databases (a terrific idea we’re researching).
By continuing to work together, we’re hopeful that the GDC will become a pivotal tool in applying precision medicine to clinical oncology and improving care for cancer patients.
Visit GDC at ASCO 2018
The GDC and other NCI experts will be at the American Society of Clinical Oncology Annual Meeting, taking place at McCormick Place in Chicago, IL. Come by to ask questions, make suggestions, or get to know DAVE!
Interactive Kiosk
June 3 & 4
9AM – 5PM
NCI Exhibit Booth #5041
Meet the Experts session: Exploring and Visualizing Data in the NCI Genomic Data Commons
Sunday, June 3
10 - 10:30AM
Presented by Michael Fitzsimons, Ph.D.
GDC User Services Manager