OCG-Supported Initiatives Provide Valuable Resources to Advance and Accelerate Precision Oncology
, by Subhashini Jagu, Ph.D. and Cindy Kyi, Ph.D.
Cancer is a global disease that calls for scientists and clinicians from around the world to join forces to advance their collective efforts. Advances in genomic studies such as high-throughput sequencing has made it possible to characterize genomes from many cancer types and learn about the genetic alterations seen in common cancers. However, the diversity of cancer subtypes and variations in individual patients’ genomes make it challenging to develop effective therapies with minimum side effects.
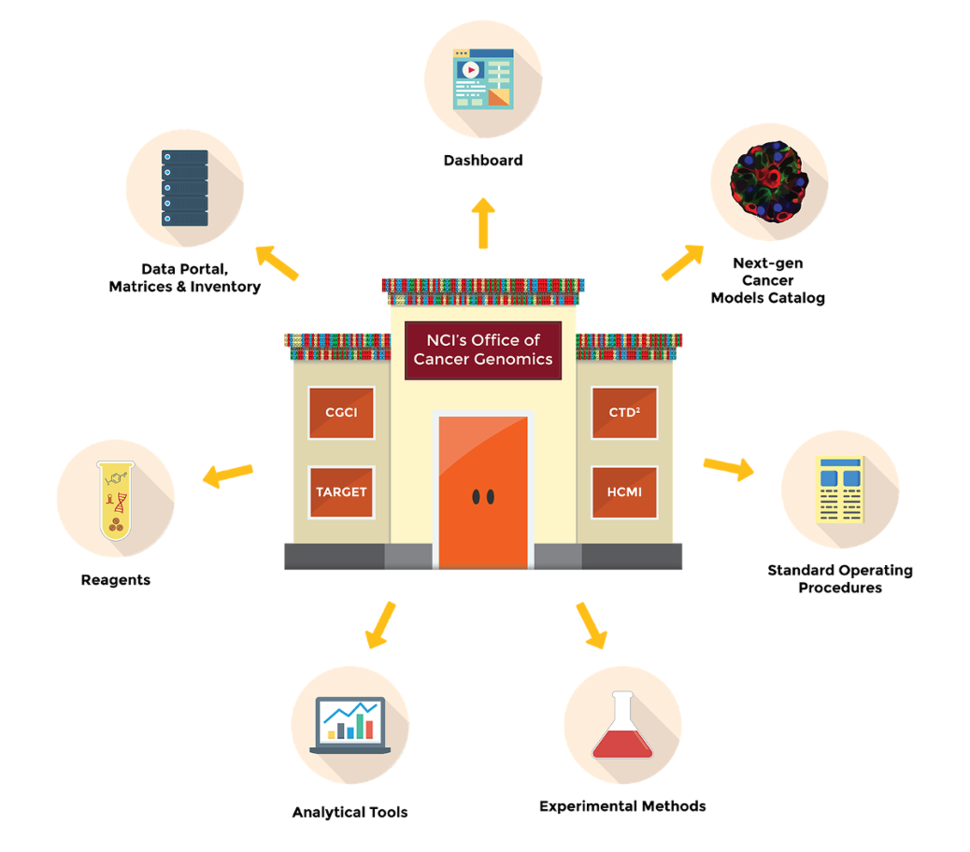
In order to alleviate some of these challenges, the Office of Cancer Genomics (OCG) supports precision oncology programs: Cancer Genome Characterization Initiative (CGCI), Cancer Target Discovery and Development (CTD2) Network, Human Cancer Models Initiative (HCMI), and Therapeutically Applicable Research To Generate Effective Treatments (TARGET). OCG initiatives share resources such as data, reagents, tools, experimental methods, and Standard Operating Procedures (SOPs) (Figure). The availability of these resources to the public fosters collaboration and encourages growth of the genomics and precision oncology fields.
OCG strongly believes that sharing data and resources will help accelerate the translational efforts towards precision oncology. The National Institutes of Health Data Sharing Policy encourages sharing of data from projects supported by public funds in a broadly accessible fashion while protecting patient’s privacy and confidentiality of proprietary data. The NCI’s Data Coordinating Center (DCC) and Genomic Data Commons (GDC) provides bioinformatics support for the OCG-supported research programs. DCC team members play a critical role in making the data available and enhancing the usability of data files by establishing the guidelines for data file formats to facilitate data mining across all projects. Thus, implementing four foundational principles—Findability, Accessibility, Interoperability, and Reusability (FAIR)—for good data management.
Accessing OCG Data
OCG initiatives share data generated through their programs with the research community in accordance with the NIH Data Sharing Policy. OCG-supported data is available in open- and controlled-access tiers. The controlled-access protects patient privacy and confidentiality. Obtaining access to controlled data and metadata files requires authorization through NCBI’s database for Genotypes and Phenotypes (dbGAP). The Data Access Guide provides the detailed instructions.
Accessing CGCI and TARGET Data
CGCI and TARGET are tumor characterization initiatives. CGCI projects use molecular characterization methods to uncover distinct molecular features of HIV+ associated cancer as well as rare adult and pediatric cancers. TARGET program utilizes comprehensive genomic characterization approach to determine the molecular changes that drive childhood cancers and to develop effective, less toxic therapies. Genomic profiles (molecular characterization and sequence data) and clinical data for a variety of tumor types are accessible through a user-friendly Data Matrix specific to each program.
Quality-controlled, standardized, raw and analyzed data from CGCI projects can be accessed through the CGCI Data Matrix ;TARGET projects can be accessed through TARGET Data Matrix and NCI’s GDC . To promote reproducible research practices, detailed experimental approaches are also provided.
Accessing CTD2 Data
CTD2 Network is a collaborative initiative that aims to bridge the knowledge gap between genomics and development of effective therapeutic strategies. Raw and analyzed primary data are available through the CTD2 Data Portal. This is a unique resource, and includes several data types such as small-molecule high-through screens, RNAi and CRISPR loss-of-function and gain-of-function screens, protein-protein interaction screens, reverse phase protein arrays etc. Researchers, who do not frequently work with large-scale, high-content data generated through high-throughtput methods should refer to “caveat emptor”.
The Network also developed the open-access web interface, “CTD2 Dashboard,” to address the community’s need to find data generated from multiple types of biological and analytical approaches by CTD2 Centers. This resource assembles Network Center-generated conclusions or “observations” with associated supporting evidence. The Dashboard allows easy navigation to biologists and data scientists.
Other Resources
Searchable Catalog of Next-Generation Cancer Models (NGCMs): HCMI is an in an international consortium that is generating patient-derived NGCMs from rare cancers and cancers from ethnic and racial minority populations. HCMI Searchable Catalog provides a list of available NGCMs to the research community. The HCMI Searchable Catalog User Guide provides instructions on how to navigate the Searchable Catalog. Harmonized genomic and clinical data of the models stored at NCI’s GDC and links to the model distributor are provided in the Catalog.
Pediatric Genomic Data Inventory (PGDI): PGDI is a catalog of known pediatric cancer projects and provides a summary of the molecular and clinical annotations.
Analytical Tools: The CTD2 Centers have developed Analytical Tools including new computational methods, algorithms, databases, and Data Portals to facilitate the processes of data mining, visualization, and analysis.
Reagents: Resources such as plasmids, cDNA clones, siRNA libraries, CRISPR-cas9 gRNA libraries, and protein-protein interaction reagents are available to the scientific community via distribution venues or by contacting the investigators that generated them.
SOPs: SOPs for current CGCI projects are provided as a reference for clinical practitioners, institutional officials, and laboratory or research personnel.
Case Report Forms (CRFs): Cancer type-specific enrollment and follow-up CRFs developed through collaborations with international clinical experts are available for use to enable uniform clinical data collection across all the tissue source sites. These forms can be used as a guide in collecting clinical data associated with cancer types.
Publications: OCG-supported initiatives have been very productive, with many manuscripts being published in high-visibility journals. In addition, most of the publications have a strong influence on the cancer genomics field and scientific community.
In summary, OCG shares valuable resources, data, and technology with the cancer research community to aid in expanding our understanding of cancer and to accelerate development of clinically useful markers, targets, and therapeutics for precision oncology. These resources are being widely used by the research community. OCG requests that users acknowledge data usage according to the guidance on relevant program-specific webpages.