HCMI Model-Associated Data Available at NCI’s Genomic Data Commons
, by Lauren Hurd, Ph.D. and Eva Tonsing-Carter, Ph.D.
Human Cancer Models Initiative (HCMI) is an international consortium whose goal is to provide a community resource of ~1000 clinically and molecularly characterized next-generation (next-gen) cancer models. The next-gen models are generated from patient tumors that span a range from common and aggressive to rare adult and pediatric cancer subtypes. OCG has partnered with NCI Center to Reduce Cancer Health Disparities to support model development from racially and ethnically diverse populations. The aim of this initiative is to provide the scientific community diverse, fully-annotated (clinical and genomics data) models which more accurately recapitulate the biology of their parent tumors.
Successful characterization of NCI HCMI models requires integration of data from multiple institutions within the NCI cancer model development pipeline. These data include NCI-supported Cancer Model Development Center (CMDC) generated clinical data collected by the Clinical Data Center (CDC), biospecimen data from the Biospecimen Processing Center (BPC) and sequencing data from the Genomic Characterization Centers (GCCs). NCI’s Genomic Data Commons (GDC) serves as a repository for NCI HCMI data, ensuring that the data is in a standardized, interoperable format. The GDC analyzes the sequencing data through specific alignment and analysis pipelines. Therefore, the data collection, analysis and sharing for use by the community is as uniform as possible.
HCMI-CMDC data at NCI’s Genomic Data Commons (GDC)
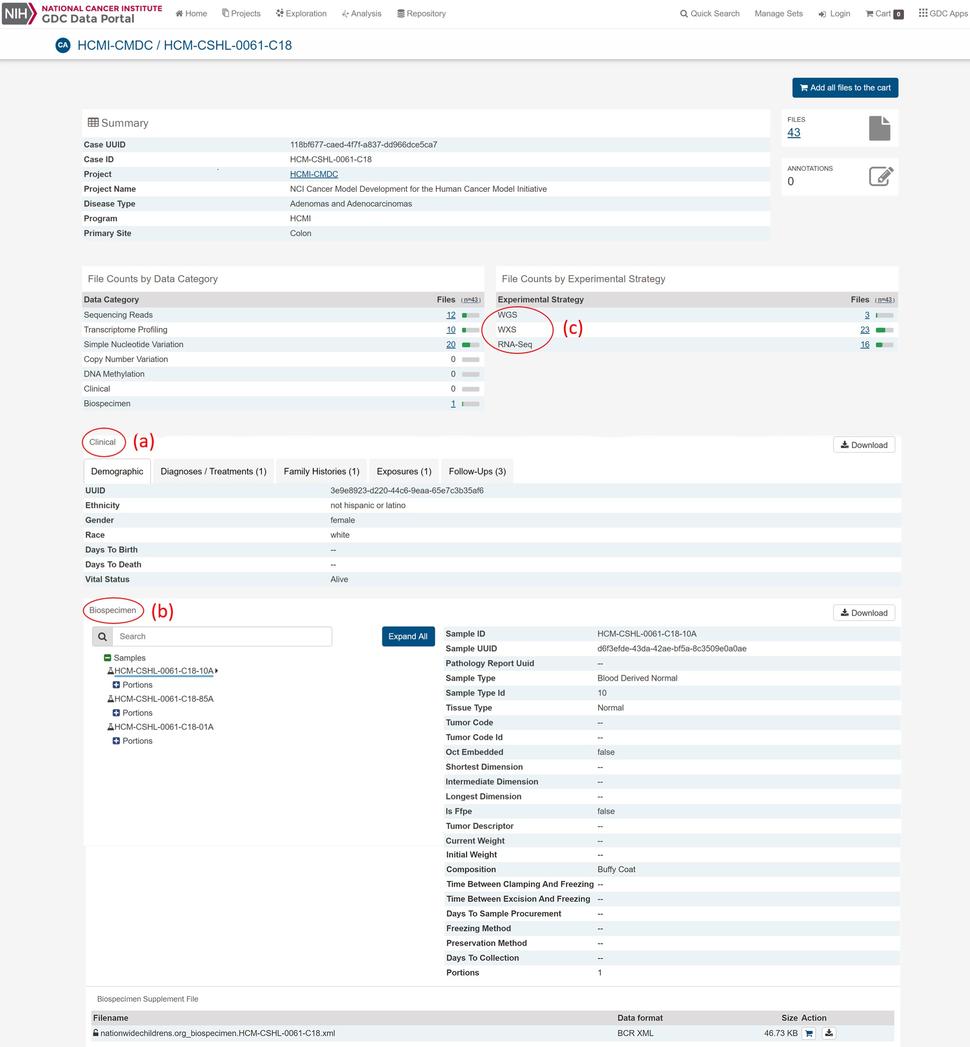
HCMI’s model-associated molecular characterization, biospecimen and clinical datasets can be accessed through GDC data portal. The GDC data portal, which also houses datasets from large-scale genomic projects such as The Cancer Genome Atlas Program (TCGA), Therapeutically Applicable Research to Generate Effective Treatments (TARGET) and the Cancer Genome Characterization Initiative (CGCI), provides users with web-based access to HCMI-CMDC and some of the data from HCMI collaborators. HCMI-CMDC data is updated as it becomes available. Currently, model-associated data are available from cancer types originating in the brain, colon, and rectum. Each model’s page details the available associated data and includes high-level clinical, biospecimen and genomic information (Figure).
The model-associated clinical data (Figure (a)) available at the GDC are submitted by the CMDCs using cancer type-specific Case Report Forms (CRFs) so that relevant diagnostic, therapeutic and prognostic information is collected. The CRFs function to standardize the clinical data and utilize clinical Common Data Elements (CDEs). The CDEs use a controlled vocabulary and are registered within NCI’s Cancer Data Standards Registry and Repository (caDSR) to ensure semantic interoperability. The clinical data are quality-controlled (QC-ed) and formatted by the CDC before the de-identified clinical data are transferred to the GDC. The GDC then harmonizes the clinical data according to GDC’s clinical data harmonization process.
Model-associated biospecimen data (Figure (b)) are also deposited at the GDC which allows for metadata collection associated with the physical sample and establishes relationships between case (e.g. model) and sample (e.g. tissue type). Prior to submission to the GDC, normal, tumor and model samples are processed by the BPC under standard protocols to ensure uniformity of nucleic acid isolation for molecular characterization. Biospecimen metadata at the GDC captures QC values for histopathology for model-associated normal and tumor tissues, such as percent tumor nuclei and necrosis, as well as nucleic acid metrics such as A260/A240 and RIN values for DNA and RNA aliquots, respectively.
Raw sequencing datasets for model and associated normal and tumor samples are submitted to the GDC by the GCCs. Nucleic acids from the BPC are sent to the GCCs where 150x whole exome sequencing (WXS) and 15x whole genome sequencing (WGS) are performed on normal, tumor and model by the DNA sequencing center and RNA-sequencing (RNA-Seq) is performed on model and tumor by the RNA sequencing center, respectively. At the GDC, the sequencing data (Figure (c)) are analyzed and harmonized using the curated GDC genomic data analysis and harmonization pipeline and ensures molecular characterization datasets are analyzed using the same bioinformatics methodology.
HCMI-CMDC datasets are either open-access or controlled. Open-access data present minimal risk that a participant can be identified and include de-identified clinical data, biospecimen data, tumor and model-associated somatic mutations. Open-access data does not require data use certification. Access to controlled datasets requires dbGaP authorization. For more information on access to controlled datasets such as raw sequencing data or harmonized datasets which contain germline variants, see the “Accessing HCMI Data” page. For accessing and downloading large sets of data, see GDC’s Data Transfer Tool.
Standardized HCMI model-associated data at NCI’s GDC available for research community
Harmonizing data through the GDC bioinformatics pipelines allows the comparison of information from multiple models or across GDC projects. GDC uses their DNA-seq analysis pipeline to identify somatic variants within the model and associated tumor from WXS and WGS data. The GDC RNA-seq pipeline is used to generate raw and normalized gene expression profiles for tumor and model data. More analytical results (e.g. alternative splicing, etc. will be added in the future). Harmonized raw sequencing data, variant calling datasets (VCFs and MAFs) and gene expression data for HCMI-CMDC models and associated normal and tumor are available at the GDC.
The results of the GDC analyses can be explored using the visualization and exploration (DAVE) tools which allows users to explore all GDC datasets including the HCMI-CMDC datasets. The GDC develops seminars and extensive documentation to orient users to the DAVE tools. Exploration pages at the GDC data portal allow for filtering of HCMI-CMDC datasets based on clinical parameters such a primary site, gender, age of diagnosis and ethnicity, etc. Once HCMI-CMDC somatic MAF files are generated, genomic data can be filtered by genes or mutations of interest. Users can select multiple model datasets and use GDC data portal tools to formulate and analyze scientific questions. Detailed information on DAVE tools and the full breath of analysis capabilities can be found at the GDC.
HCMI’s objective is to provide the research community with a resource of next-generation cancer models that are characterized with clinical and molecular data. Providing datasets in a standardized and unified manner, in alignment with four foundational principles—Findability, Accessibility, Interoperability, and Reusability (FAIR), is essential for efficient data sharing and enables users to compare multiple datasets. NCI’s GDC plays an integral role in helping HCMI meet this objective through its systematic and harmonized data processing, organization, storage and analysis tools. Users may join the GDC User Mailing List to receive updates on data releases.