Human Cancer Models Initiative’s Searchable Catalog of Cancer Models
, by Cindy Kyi, Ph.D., Eva Tonsing-Carter, Ph.D., and Lauren Hurd, Ph.D.
HCMI Background
The National Cancer Institute’s (NCI) director, Dr. Ned Sharpless, has made a commitment to focus on efforts in “big data” and build databases as common resources for researchers to accelerate the translation of laboratory findings into the clinic. In harmony with this focus, a goal of the Office of Cancer Genomics (OCG) is to facilitate the development and sharing of resources such as data, tools and protocols within the research community.
The Human Cancer Models Initiative (HCMI), one of OCG’s programs, is an international collaboration founded by the NCI, Cancer Research UK (CRUK), Wellcome Sanger Institute (WSI), and the foundation Hubrecht Organoid Technology (HUB). The ultimate goal of HCMI is to provide a community resource of patient-derived next-generation cancer models (e.g. 3D organoids, 2D conditionally reprogrammed cells, etc.) with associated molecular sequencing and clinical data. The motivation behind HCMI’s next-gen cancer models is discussed in this previous e-News article. The models, as well as protocols and model-associated data, are available for the research community to be used in basic and translational research.
NCI-funded cancer model development centers (CMDCs) and HCMI consortium members are creating about 1,000 models from diverse tumor types including rare, pediatric cancers, and cancers from racially and ethnically diverse populations. HCMI models are annotated with patients’ clinical data, the genomes and transcriptomes of the derived model and associated parent tumor and matched normal tissue from most cases. Molecularly characterized sequences and associated clinical data for the models from the NCI-supported CMDCs will be available to researchers through NCI’s Genomic Data Commons (GDC). The HCMI models will be available to researchers through the third-party distributor, American Type Culture Collection (ATCC).
For the detailed process of NCI cancer model development pipeline, please see the NCI Cancer Model Development page. Once the HCMI cancer model development pipeline is completed, the validated models, clinical and molecular data, and tools are available as a community resource.
As an ongoing community resource, HCMI is developing an online interactive Searchable Catalog for potential users to view the available models and associated information in a centralized place. In this catalog, users can query, select, view, and download available information regarding the HCMI models.
HCMI Searchable Model Catalog
The HCMI Searchable Catalog is a result of joint efforts and collaboration between all members within the HCMI. The HCMI Searchable Catalog contains data elements extracted from the quality- checked clinical and molecular characterization data for each tumor and derived model. The data elements were developed and compiled with input from clinical collaborators and approved by the HCMI steering committee members. The data elements are defined by controlled vocabulary through collaboration with staff from NCI’s Cancer Data Standards Registry and Repository (caDSR). Additional data elements will be added in the future and feedback from users will be taken into consideration.
Browsing the Searchable Catalog Landing Page
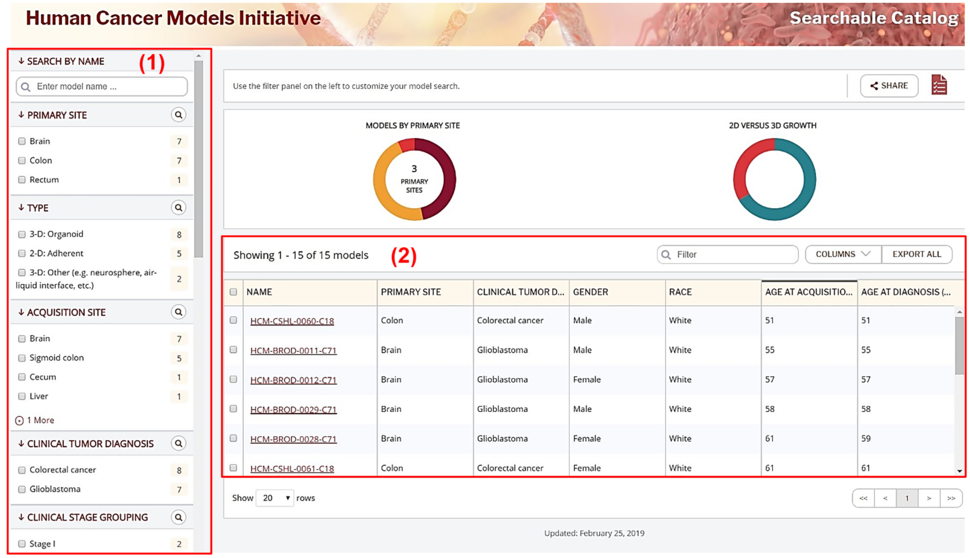
The catalog allows users to filter and identify models according to data element of interest. The filter panel (1) on the catalog landing page (Figure 1) displays the searchable data elements, which include characteristics of the models such as the primary site of tumor, type of models, tissue acquisition site as well as the clinical diagnosis and background of the patients that the models were derived from. The models that meet the characteristics chosen in the filter panel are displayed in the main display pane (2) on the landing page. Users can choose the data element of interest under “COLUMNS” to be displayed within the main display pane. The catalog also allows users to export the model information displayed in the main viewing panel by using the “EXPORT ALL” function. Users may also save models to “My Model List” by checking the box next to the model name. “My Model List” models and associated data can be downloaded by clicking the “Download TSV” tab within “My Model List”.
Browsing Individual Model Pages
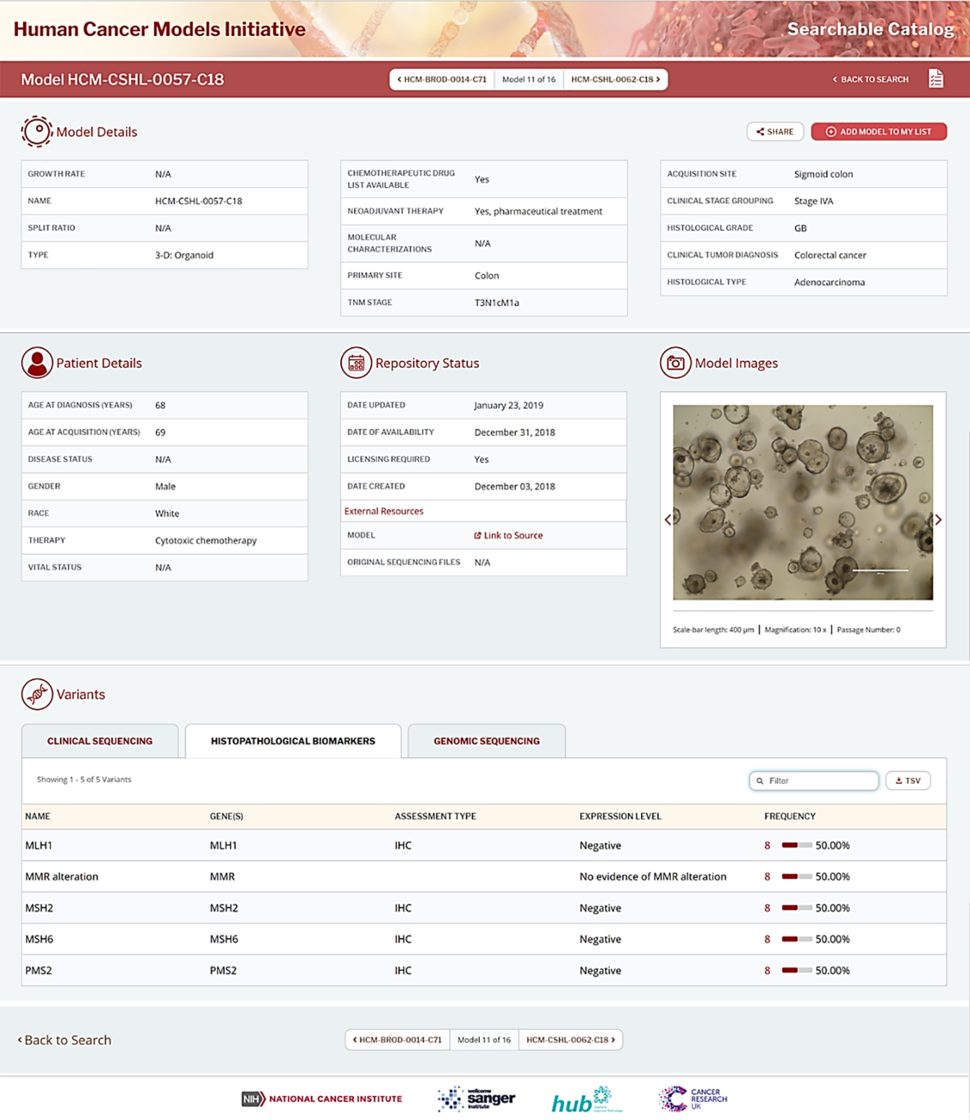
Users may view detailed information on individual models by clicking on a specific model name under the “NAME” column in the main display pane (Figure 1). Clicking on an individual model name will take users to the model page (Figure 2) with more detailed information about that model.
Under “Model Details”, users can find details of the model such as growth rate, split ratio, and the type of model as well as tumor-related information such as therapeutic regimen, primary site of cancer, stage of cancer (TNM stage), etc. In addition, “Patient Details” include demographics and clinical background of the patient from whom the tissue sample for the model was acquired. Furthermore, under the “Repository Status”, users can find information about licensing requirements for commercial use, availability of the model at the third-party distributor, and links to external resources including NCI’s GDC website. At the GDC, users can find available associated clinical and sequencing data of the parent tumor, matched normal tissue, and the model. Users who are interested in purchasing HCMI models for use in research may also follow the link to the third-party distributor, ATCC.
Users may view the images of growing cells from a specific model under “Model Images” at different magnifications as available. The cell growth pattern on images will vary according to the model type (e.g. 2D or 3D).
The “Variants” section on the model page includes information on “clinical sequencing”, which shows any collected clinically-derived sequence variants, clinical “histopathological biomarkers” of the cancer as well as information pertaining to each biomarker, and the “genomic sequencing” of the models and associated tissues as available. Users may also enter keywords in the “Filter” box to filter for specific variants (e.g. MSH6, TP53, etc.). Variant data for the model can be downloaded by clicking on the “download TSV” icon.
For more detailed Searchable Catalog information, users may see the HCMI Searchable Catalog User Guide.
HCMI’s Continuous Efforts Towards Precision Oncology
HCMI is generating models from breast, colorectal, esophageal, glioblastoma, liver, and pancreas. Models for rare pediatric cancers such as neuroblastoma, Wilms tumor, and Ewing sarcoma are in progress. HCMI also plans to develop models from other cancer types including ovarian, head and neck, kidney, and bladder as well as cancers from racial and ethnic minority populations. The model list will be updated as the models become available.
The aspiration for HCMI is to provide a resource of novel next-generation cancer models that are characterized with clinical and molecular data to the research community. HCMI models and resources such as protocols used for model development and SOPs may provide the research community with the tools to study the molecular pathways that influence tumor development in these cancers and contribute to precision oncology.