Leveraging a Genomics Background to Facilitate Molecular Characterization of HCMI Models
, by Lauren Hurd, Ph.D.
My name is Lauren Hurd and I am a Scientific Program Manager for the Human Cancer Models Initiative (HCMI) within the Office of Cancer Genomics (OCG). National Cancer Institute (NCI), together with other consortium members, co-founded HCMI to provide a resource of ~1,000 clinically and molecularly characterized next-generation patient-derived cancer models to the research community. The cancer models are generated from parent tumors that span a range of subtypes originating from individuals of diverse ethnic and racial backgrounds as well as rare adult and pediatric tumors.
The goal of this initiative is to provide a resource of diverse, fully annotated models which more accurately recapitulate the biology of their parent tumors. As a scientific program manager, I work with Daniela Gerhard, the Director of OCG to ensure that this goal can be met for the United States' (U.S.) contribution to the HCMI.
For the last eight years, I have been working within the field of genomics in dynamic and challenging roles. My interest in genomics was cultivated in graduate school, where I focused on understanding the relationship between pathogenic variants in a nonselective cation ion channel and an autosomal dominant form of skeletal dysplasia, a disorder that affects the growth of bone and cartilage. I continued my career in genomics during my postdoctoral fellowship where I led the technical direction of molecular diagnostic testing for rare pediatric disorders in a clinical sequencing lab. It was in this role that I gained extensive knowledge of both genomic testing and genomic data. I developed Sanger and NGS panel testing from the ground up, interpreted variants identified during testing and reported on clinically actionable results. Meaningful interpretation or curation of the variants identified through the various sequencing technologies quickly became one of my favorite parts of the job.
Prior to joining NCI, I managed a team of variant curation scientists who were responsible for interpreting variants identified during routine carrier screening of autosomal recessive and X-Linked pediatric disorders. I also collaborated with a multi-disciplinary team from bioinformatics, marketing and clinical operations to develop new versions of the carrier screening panel which required critical management of many deliverables on strict timelines.
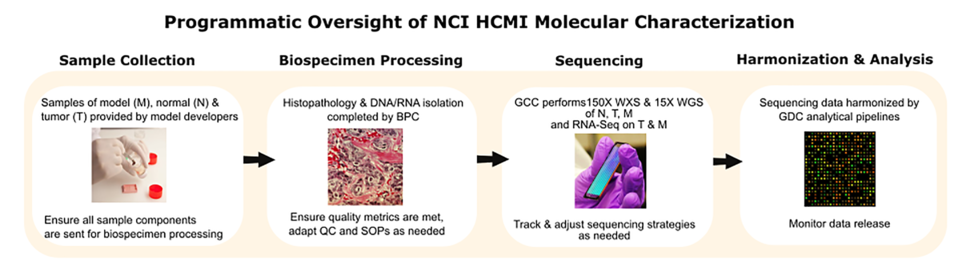
HCMI models within the U.S. are molecularly characterized with genomic and transcriptomic data for the model as well as the associated normal, and parent-tumor. Annotating the models with this data, to the best of our ability, is a complex process. I leverage my strong background in molecular biology and genetics as well as my experience in project management to facilitate this process (Figure). I work closely with the Biospecimen Processing Center (BPC) to ensure that quality nucleic acid samples from model, normal and parent-tumor can be provided for downstream sequencing to the Genomic Characterization Centers (GCCs) and subsequently harmonized by the Genomic Data Commons (GDC). This involves monitoring hundreds of samples through the molecular characterization pipeline, tracking histopathology and nucleic acid metrics, and ensuring sequencing strategies are adjusted as needed.
I also collaborate with these teams to identify potential challenges in the molecular characterization process and develop resources which address those challenges. I’ve developed standard operating procedures (SOPs) for the Data Coordinating Center (DCC) to increase the interoperability of model QC data and the BPC to streamline the nucleic acid isolation process. HCMI models are most valuable when they contain comprehensive and standardized datasets and I do the best I can to ensure that we provide these datasets to the research community.
While I am extremely familiar with pediatric and rare disease genomics, the field of cancer genomics is mostly uncharted territory for me. The HCMI program has provided a whole new perspective on learning about genomics. Providing models annotated with clinical, genomic and transcriptomic datasets provides researchers with a crucial resource to address large questions within the field.
Study of the models will advance our basic understanding of tumor heterogeneity, genomic stability, tumor-immune microenvironment and mutational signatures on a more comprehensive scale. Large repositories of molecularly diverse models from the same tumor subtype will also allow for greater representation when screening for novel therapeutic targets as well as assessing drug sensitivities. All of this will certainly culminate in the advancement of both novel and improved precision therapies. It is exciting to be a part of this initiative where the possibilities for discovery seem endless.
The human nuclear genome consists of approximately 3.2 billion nucleotides. An extraordinary amount of biological information is brought together both in precise order and time to form the foundation of human life. It’s fascinating and logical, yet puzzling, all at the same time. It’s certainly what has driven me to pursue a career in genomics. There are so many ways in which one can work in the field of genomics and I have been fortunate to work in this field in many capacities. While the career path I have taken is varied in its relationship to the field of genomics, one commonality remains. I work with teams and initiatives who excel at providing answers to the greater scientific community and the HCMI is no exception.