GenePattern Notebook: Integration of Electronic Notebooks with Bioinformatics Tools for Genomic Data Analysis
, by Michael Reich
Over the past several years, the electronic analysis notebook has emerged as an effective and versatile tool for the authoring, publishing, and sharing of scientific research. It allows scientists to combine the scientific exposition – text, images, and even multimedia – with the actual code that runs the analysis, creating a single “research narrative” document that is reproducible, containing all of the computational steps in an analysis; adaptable by other scientists to their own research; comprehensive, conveying research in a high level of detail, without the limitations of publications or paper media; and accessible, often requiring only a web browser to view and run.
The Jupyter Notebook system1 has become a de facto standard notebook environment in data science and genomic analysis. The community of Jupyter users extends well beyond these, reaching areas of science as diverse as physics, economics, and linguistics. However, the Jupyter notebook format assumes familiarity with a programming language in order to access analyses, and even text must be formatted using a programming-style language.
To extend the capabilities of notebooks to the needs of researchers at all levels of programming expertise, we developed the GenePattern Notebook environment2 with funding from the National Cancer Institute’s Informatics Technologies in Cancer Research (ITCR) program. GenePattern Notebook (http://genepattern-notebook.org/) integrates Jupyter’s research narrative capabilities with the hundreds of genomic analysis and visualization tools available through the GenePattern platform.3 The GenePattern Notebook workspace is available for public use and requires registration to gain access. This tool allows scientists to develop, share, collaborate on, and publish their notebooks, requiring only a web browser. In this environment, investigators can design their in-silico experiments, perform and refine analyses, launch compute-intensive analyses on cloud-based and high-performance compute resources, and publish their results as electronic notebooks that other scientists can adopt to reproduce the original analyses and modify for their own work.
The GenePattern Notebook environment provides capabilities beyond standard notebook platforms:
Access to a wide range of genomic analyses within a notebook
GenePattern provides hundreds of analyses, from machine learning techniques such as clustering, classification, and dimension reduction, to omic-specific methods for gene expression analysis, proteomics, flow cytometry, sequence variation analysis, pathway analysis, and others. The analyses are launched from a user-friendly form in the notebook and run on a remote GenePattern server, which may be on a cloud provider or hosted at a high-performance compute site. This allows compute-intensive analyses to run in an environment where they are most suited. Results are available within the notebook and may easily be used in other analysis steps.
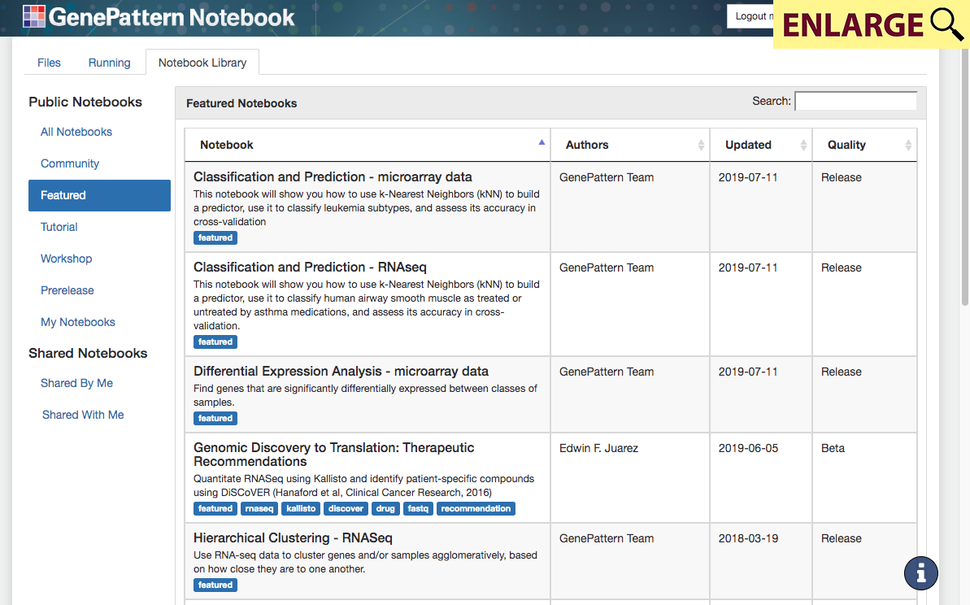
Featured notebooks
A library of featured genomic analysis notebooks is available on the GenePattern Notebook workspace (Figure 1). These notebooks include templates for common analysis tasks (e.g. hierarchical clustering of RNA-seq data, gene set enrichment analysis, non-negative matrix factorization (NMF)), as well as disease-specific research scenarios and compute-intensive methods. Featured notebooks also include those that were developed in collaboration with research labs as a means to disseminate their analysis methods, including the Coordinated Gene Activity in Pattern Sets (CoGAPS) Bayesian NMF method for inference of biological process activity4 and the AMARETTO multi-omics tool for inference of regulatory networks in cancer and other diseases.5 A cancer-focused notebook, “Genomic Discovery to Translation”, is aimed at providing insights into candidate drugs for patient therapy. This method combines RNA-Seq profiling data with expression and viability data from cell lines to identify compounds as candidate therapeutics. It uses publicly available data resources, including the Cancer Cell Line Encyclopedia (CCLE), Sanger Cell Line Project (SCLP), Cancer Therapeutics Response Portal (CTRP), and Genomics of Drug Sensitivity in Cancer (GDSC).
Scientists can easily copy these notebooks, use them as is, or adapt them for their research purposes. Users with computational experience can modify their own versions of the notebook with variations, for example to try alternative analysis methods, additional data resources, or other ‘omics’ data types. An example notebook for the analysis of copy number variation in methylation array data is shown in Figure 2. The GenePattern analysis shown there replaces a considerable amount of code and facilitates analysis for non-programming scientists. Researchers can upload and store up to 30 GB of data and GenePattern development team can increase the size if additional space is required for the analysis.
Notebook enhancements
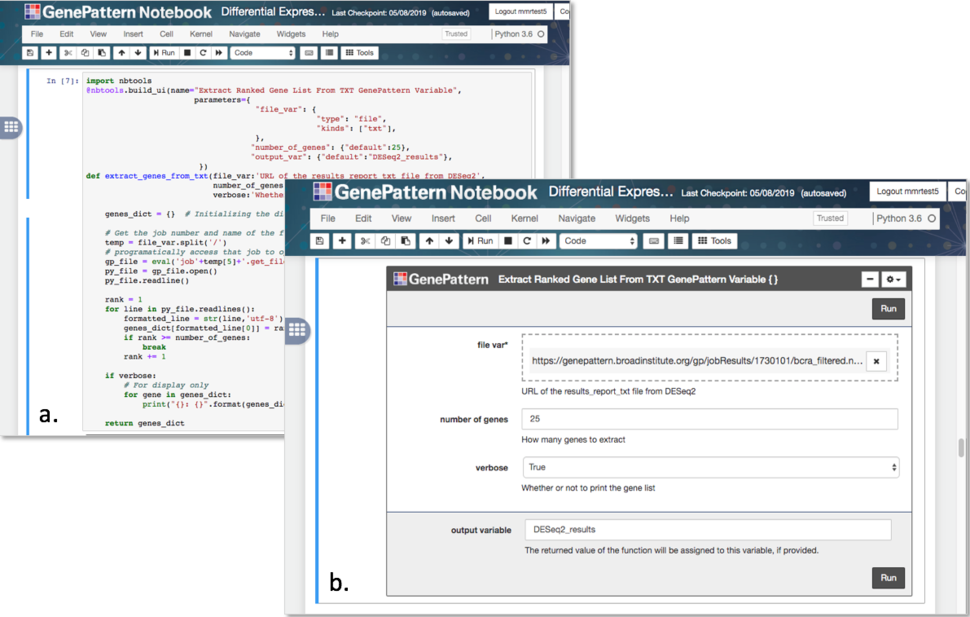
GenePattern Notebooks have several features that enhance the original standard Jupyter notebook interface. First, a rich text editor allows scientists to enter and format text without knowing a text formatting language such as Markdown or LaTeX. Second, users can create a table of contents from the headings in a notebook, which updates automatically as headings are added or changed. It can either be embedded in the notebook or float alongside, allowing easy navigation to any point in a notebook. Third, a user interface-building tool (Figure 3) allows notebook developers to wrap their code so that it is displayed as a web form, with only the necessary inputs exposed. Users of the notebook are presented with a simplified display that allows them to run the analyses without needing to interact with the code behind them.
Publication and collaborative editing
Notebook developers often wish to share their notebooks, either with the research community or among collaborators. To make a notebook publicly accessible, the author selects the “publish” feature and adds descriptive information and tags to make the notebook easy to find in a search query. The notebook is then made available on the “community” section of the workspace. An author can include a web link to a public notebook in a publication, and users who follow the link will see a read-only version of the notebook, with the option to log in to the workspace, where they can run, copy, and edit their own version. For collaborative editing, an author can send a sharing invitation to colleagues, who then can also view, run, and edit the notebook prior to its publication.
The GenePattern Notebook environment is freely available at http://genepattern-notebook.org/. Researchers can make their tools available for public use through the GenePattern server or GenePattern Archive (GParc), a community repository. A related GenePattern Notebook resource, the Human Cell Atlas Notebook Workspace, https://hca.genepattern.org/, is dedicated to the Human Cell Atlas6 and features a growing collection of notebooks providing single-cell analysis tools. For more details about the GenePattern Notebook, view the video tutorial at https://genepattern-notebook.org/tutorials/.
References
- Kluyver T, Ragan-Kelley B, Pérez F, et al. Jupyter Notebooks-a publishing format for reproducible computational workflows. In ELPUB. 2016 May 26; pp. 87-90.
- Reich M, Tabor T, Liefeld T, et al. The GenePattern Notebook Environment. Cell Systems. 2017 Aug 23;5(2):149-151.e1. (PMID: 28822753)
- Reich M, Liefeld T, Gould J, et al. GenePattern 2.0. Nature Genetics. 2006 May;38(5):500-1. (PMID: 16642009)
- Fertig EJ, Ding J, Favorov AV, et al. CoGAPS: an R/C++ package to identify patterns and biological process activity in transcriptomic data. Bioinformatics. 2010 Nov 1;26(21):2792-2793. (PMID: 20810601)
- Champion M, Brennan K, Croonenborghs T, et al. Module Analysis Captures Pancancer Genetically and Epigenetically Deregulated Cancer Driver Genes for Smoking and Antiviral Response. EBioMedicine. 2018 Jan;27:156-166. (PMID: 29331675)
- Regev A, Teichmann SA, Lander ES, et al. The Human Cell Atlas. Elife. 2017 Dec 5;6. pii: e27041. (PMID: 29206104)