Seeing Cancer Through New Lenses: A Broader Spectrum of Cancer Targets
, by Roya Rafiee, M.Sc., Ph.D.
For over 17 years, Office of Cancer Genomics Director Dr. Daniela Gerhard led a great number of large-scale genomics research efforts at NCI. Dr. Gerhard had a special knack for imagining how genomics could change research technology and shape how we understand and study cancer.
Dr. Gerhard very sadly and suddenly passed away last June. Among the many programs Dr. Gerhard was running at the time was The Cancer Target Discovery and Development (CTD2) program, focused on translating the abundance of genomic characterization data into functional insights about cancer, and in turn, new treatments.
One of her most recent publications describes the current landscape of functional genomics research—new ways to think about cancer and its vulnerabilities that will give rise to much needed, new treatments for patients. Improving the lives of patients was truly what motivated Dr. Gerhard in all her work—and did she ever work hard! Though she has passed, her many programs will continue with this same motivation.
- Dr. Louis Staudt, Center for Cancer Genomics Director
Despite much progress in genomics and medicine, most patients do not have a targeted therapy available for their specific disease and cancer still causes millions of deaths every year.
Though identifying therapeutic targets for cancer has been a high priority for some time, few therapies have shown efficacy in clinical trials. Attempts to target somatically altered oncoproteins have been successful. For example, the robustness of BCR-ABL as a driver oncogene in chronic myeloid leukemia (CML) has made imatinib a game-changing drug for the disease.1 However, most cancers fail to express BCR-ABL or other targets with existing therapies.
A more fruitful tactic may be to focus on identifying the “non-oncogene” dependencies of cancers, i.e., genes that are not involved in the development of the tumor but are part of normal cellular functions necessary for the ongoing maintenance and survival of the tumor. Many of these genes are rate-limiting components of their pathway, making them ideal drug targets.2,3 Emerging research suggests that the variety of potential therapeutic targets resulting from non-oncogene dependencies is much broader than the list of frequently mutated genes.3
“The ongoing discovery of non-oncogene targets represents an important opportunity to develop new approaches against cancers,” says William Hahn, M.D., Ph.D. of the Dana-Farber Cancer Institute.
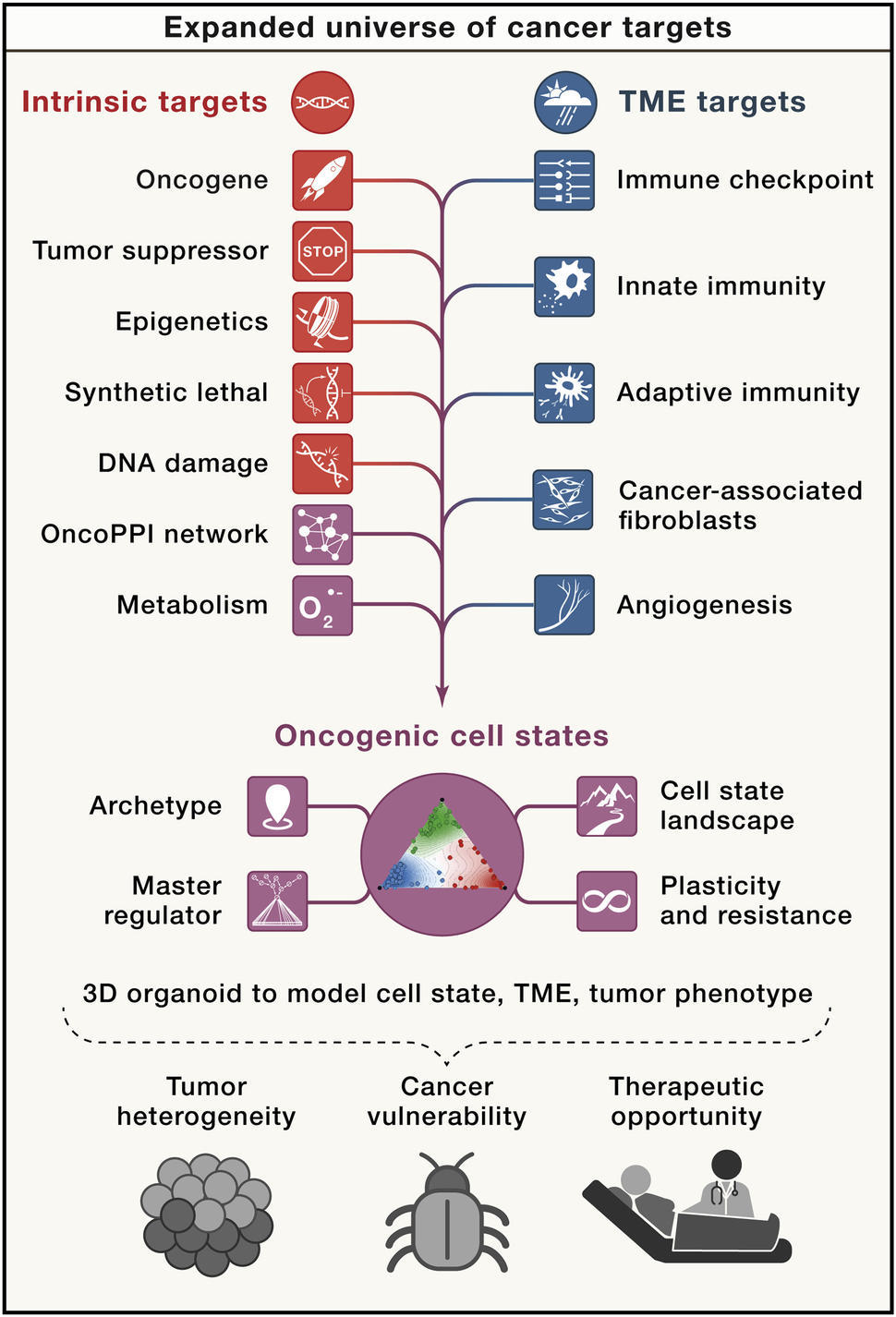
In “An expanded universe of cancer targets” published recently in Cell, Dr. Hahn and other members of NCI’s Cancer Target Discovery and Development (CTD2) Network discuss how considering new classes of cancer targets outside of the proverbial oncogene may accelerate the discovery of therapeutic agents.3
Thinking Outside the Tumor Box
Targets may be divided into either intrinsic (not dependent upon interactions with other cells), or extrinsic (tumor microenvironment-mediated) to the tumor (see figure).3 Intrinsic factors include mutations of oncogenes, epigenetic changes, dysregulations of transcription and signal transduction, abnormal metabolic responses, and altered DNA damage responses (DDRs). In some cases, when the signaling induced by oncogenes is eliminated, cancer cells that harbor these oncogenes are prone to cell death, a circumstance known as oncogene addiction.2
A variety of tumor-extrinsic factors also contribute to tumorigenesis, including changes in the tumor microenvironment (TME), alterations to the differentiation status, and composition of tumor-infiltrating stromal cells, such as immune cells and fibroblasts. Erroneous endocrine signals and the microbiome can also play an important role in cancer pathogenesis and its response to treatment. The ability for a cell to change their identity, differentiation status, or other phenotype in response to extrinsic factors—a concept known as plasticity—may also help tumors evade therapies.3
Modeling Tumor Cells with Discrete States
The concept of oncogenic “cell states,” which model subcellular properties in tumors such as drug sensitivity, metabolism, and stemness, may help enable better understanding of the complexity of a tumor. A single tumor at one moment in time is composed of cells in many distinct cell states, even if the cells are isogenic.3
Recent studies are considering how the length of time a cell stays in a particular state (generally categorized to as “stable” or “meta-stable”) and the transitions between states may affect both tumor progression and sensitivity to therapeutic agents.4,5 Some studies suggest that the presence of specific cell states and transitional states may affect disease progression more than other factors such as tumor histology or genetics. E.g., a mixed epithelial-mesenchymal state has been linked with poor outcomes in breast cancer patients.6 Considering cell states may give rise to new therapies that address tumor heterogeneity and are independent of oncogene expression.7
Decoding and Targeting the Tumor Microenvironment
It is crucial that cancer be targeted not in isolation, but rather in the context of a larger ecosystem to improve patient outcomes. Although the complex composition and activities of the TME are still unsolved, notable clinical success utilizing TME information already exist in immune- and blood vessel-targeted interventions.8
Though targeting a tumor’s surrounding microenvironment may be key piece in fully eradicating a tumor, it is imperative to keep in mind the vital functions that tissue microenvironment components serve in normal contexts. E.g., therapeutic targeting of tumor stroma may result in toxicity to healthy stroma elsewhere in the body. Here, focusing treatment on a particular oncogenic cell state could simplify and guide the development of therapeutics and reduce toxicity.
“These efforts will also facilitate the development of combination therapies that combine different targets and vulnerabilities in cancers,” says Dr. Hahn. A robust method for evaluating combinations of therapies will also be needed in the future, particularly if targeting intrinsic and extrinsic factors together. Developing these types of approaches will require new ways to evaluate cell fitness and means to measure tumor viability. Utilizing different approaches to modeling cancer and the complex TME may identify new kinds of targets and rational combinations that provide robust, durable, and effective treatment for patients.9,10