EcDNAs are Extrachromosomal, Extra Active, and Extra Terrifying: Researcher Interviews with Chia-Lin Wei, Roel Verhaak, and Howard Chang
, by Peggy I. Wang
Extrachromosomal circular DNAs (ecDNAs), or particles of DNA existing outside the autosomal genome, were discovered in the 1960s and more recently have been implicated in cancer development.
EcDNAs frequently occur across many cancer types and often in high copy numbers. The oncogenes they carry are thought to be highly expressed compared to copy number-matched linear DNA. Cancers carrying ecDNAs are also associated with shorter survival for patients.
I spoke with several researchers who are looking into distinct mechanisms for how exactly ecDNAs dysregulate gene expression and contribute to oncogenesis: Dr. Chia-Lin Wei and Dr. Roel Verhaak, professors at the Jackson Laboratory who examined the interactions ecDNAs may have with chromatin, and Dr. Howard Y. Chang, who examined the interactions ecDNAs may have with each other. Their complementary models of ecDNA function point to an overall major role for ecDNAs in cancers. A better understanding of ecDNAs may soon reshape our understanding of how cancer develops and evolves and also provide insights into therapeutic strategies.
Peggy I Wang: What are extrachromosomal circular DNAs (ecDNAs)? Do they have any known “normal” functions?
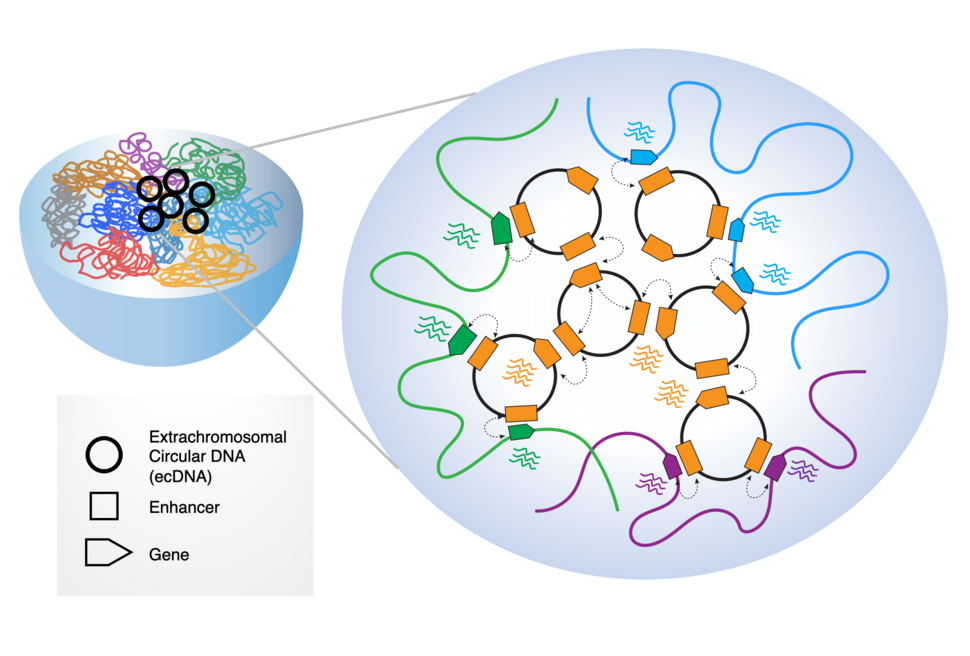
Roel Verhaak: Extrachromosomal circular DNA elements are pieces of DNA that have broken off the linear chromosomes and circularized. There are two types: small 100 bp – 10 kb elements that can be found in many different cell types in the body, with unknown function. And then there are larger (50 kb – 5 Mb) oncogenic elements, which are only detected in cancer cells and carry genes known to activate cancer cells. These oncogenic ecDNAs are found in ~15% of newly diagnosed cancer.
PIW: Could you describe the different types of chromatin interaction assays you've used to study ecDNAs, such as ChIA-PET and ChIA-Drop? What kind of different information do they provide?
Chia-Lin Wei: These are methods used to map genome-wide, long-range chromatin interactions between regulatory elements. ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag sequencing) is a method we developed to reveal the general spatial chromatin organization and to identify chromatin interactions associated with specific proteins. The resulting paired sequences from ChiA-PET tell us about the connectivity between different genomic regions and the 3D organization of the chromatin.
While informative, ChIA-PET is limited in that it is only telling us about pairwise interactions aggregated from bulk cells—we don’t know whether or not those pairs are occurring together within a single complex. So we have developed the ChIA-Drop (Chromatin Interaction Analysis by Droplet sequencing) chromatin interaction method to identify the combinations of chromatin interactions that occur within a single complex.
PIW: What did you find when you applied these chromatin interaction assays to cell lines known to have ecDNAs?
CLW: Our goal was to determine the spatial chromatin organization and chromatin interactions of ecDNAs in general. Given the circular structure of ecDNAs, we anticipated that they would exhibit unique spatial patterns.
We uncovered very high levels of chromosome connectivity and transcriptional activity: the ecDNAs exhibit a pattern of dense and widespread chromatin interactions with actively transcribed genes and regulatory elements that reside both within the ecDNA (in cis) and on the chromosomes (in trans).
We reasoned that this is because the small size of ecDNAs allow them to move freely amongst the chromosomes. Such mobility could enable ecDNAs to interact with genes residing on chromosomes. Moreover, the interaction sites on the ecDNAs exhibited key characteristics of super-enhancers (SEs), which are known to exert a unique regulatory influence that could promote tumorigenesis.
RV: With its mobility and potentially high copy numbers, ecDNAs can potentially transverse the nucleus, and function as trans-acting, mobile transcriptional enhancers, establishing extensive chromosomal interactions and driving transcription of specific chromosomal genes. Thus ecDNAs may be a very powerful mechanism to promote cellular fitness.
PIW: So you found that ecDNAs have more interactions genome-wide than previously understood. Is that where the nTIF metric comes in?
CLW: The nTIF metric, we synthesized to remove copy-number bias in the detection of ecDNA–chromosome interaction frequencies and quantitatively measure normalized trans-interaction frequency between regions from different chromosomes or between ecDNAs and chromosomes. Two regions with higher a nTIF value indicate higher connectivity.
We observed a sharp elevation of nTIFs in ecDNA regions compared to the interaction frequencies of their corresponding native chromosomal regions in cells without ecDNAs. These interactions were specifically enriched with chromosomal promoters. Chromosomal genes whose promoters interact with ecDNAs were expressed at significantly higher levels.
PIW: Is it difficult to distinguish interactions between a chromosomal region and the chromosomal copy of an oncogene, versus an ecDNA copy of the gene?
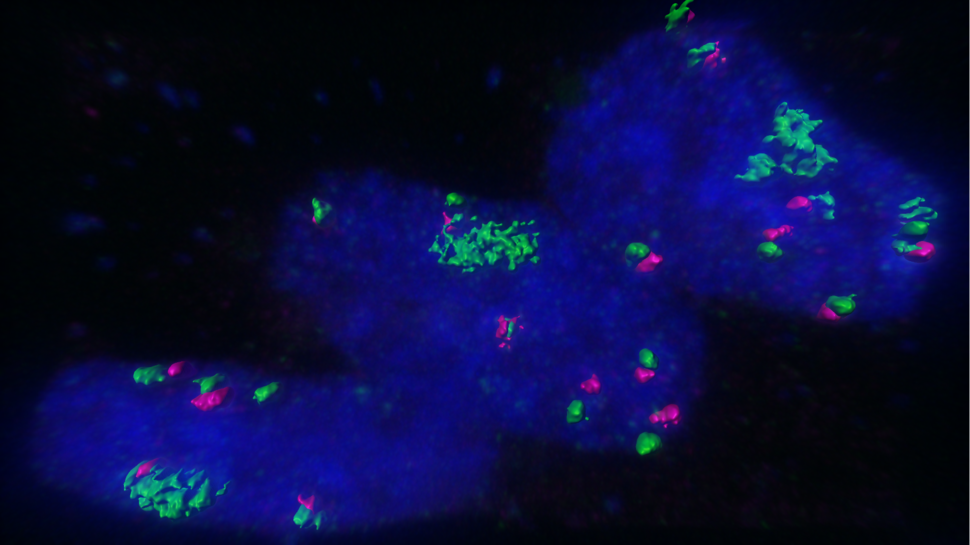
CLW: It is really challenging to accurately assign chromatin interactions to ecDNAs or chromosomes, given their nearly identical sequences. However, since ecDNA copy numbers far exceed chromosomal copy numbers, and chromosomal interactions are known to be greatly physically limited by their organization and structure, we believe that the majority of interactions we found are contributed by ecDNAs. In our study, we also perform imaging-based validation of interactions between ecDNAs and specific chromosomal sites.
PIW: In addition to interacting with chromatin, another working model for ecDNA oncogenic activity is that ecDNAs are interacting with each other, in something called ecDNA “hubs.” Can you describe more about what these hubs are?
Howard Y. Chang: EcDNA hubs are basically micron-sized collections of 10 to 100 copies or more of ecDNA molecules. We discovered that these collections appear to be a prevalent form of ecDNA occurrence in cancer cells and basically a site for massive oncogene transcription.
PIW: And what is unique about how the ecDNAs you studied activate oncogenes?
HYC: In the normal chromosomal context, to turn on a protein coding gene, the cell needs enhancers and promoters to control when the gene turns on and off. And those control elements are nearly always located on the same chromosome.
We discovered that inside the ecDNA hub there's very promiscuous sharing of DNA regulatory elements—one ecDNA can actually use enhancers from other ecDNAs, even if they derive from originally different chromosomes! For example, a gastric cancer we studied contains an ecDNA containing the MYC oncogene coming from chromosome 8 and also a separate ecDNA with FGFR2 from chromosome 10. When these ecDNAs intermix, an ecDNA oncogene can get all this input from other molecules, resulting in a very strong level of transcription. Using an enhancer from one ecDNA to activate an oncogene on another ecDNA is what we mean by "intermolecular enhancer–gene interactions."
PIW: How did you discover that the enhancers are being shared from different ecDNA molecules?
HYC: We used CRISPR interference to silence individual enhancers and looked at the effect on oncogene expression on different ecDNAs. And we in fact found that the enhancers situated on different molecules activated the oncogenes.
PIW: Are the individual ecDNAs working together in a coordinated fashion to transcribe oncogenes?
HYC: We indeed found what could be described as “cooperative expression.” When we measured, for every molecule of ecDNA, how likely it is to make an RNA product, we discovered that each ecDNA is more likely to fire and transcribe when it's near in physical proximity to other ecDNAs. So each ecDNA molecule is not doing their own thing or stochastically turning on and off. They help each other transcribe. When they're in these hubs, each one of them is more likely to actually turn on the oncogene product.
PIW: It seems so easy for an ecDNA to just ramp up oncogene expression (through either model)! Should we be terrified?
HYC: It really is kind of terrifying—we often think about the protein products of oncogenes as what is helping the cell. But here, we’re talking about the oncogenic DNA itself affecting each other’s gene expression. Going back to this phenomenon of intermolecular activation that I mentioned earlier with gastric cancer example: once both MYC and FGFR2 become ecDNAs, they intermix, and now DNA elements from one is turning on the oncogene on the other one and vice versa. So normally those two sets of genes don't talk to each other or see each other, but now they have a chance to get together and have this very nefarious mode of gene activation.
PIW: Are your experiments with the small molecule JQ1 a potential therapeutic strategy against ecDNAs?
HYC: Using a 3D genome conformation assay called HiChIP, we found high levels of intermolecular enhancer-promoter contact. And we think the biochemical mechanisms for enhancer-promoter contact is what's holding the ecDNA hubs together.
So we were actually testing the idea that the ecDNA hub is held by is held together by a protein core—in this case a BET protein called BRD4. If you block the function of that protein with a BET inhibitor, the ecDNA molecules fly apart, the hub gets broken up, these DNA molecules fly apart and the transcription stops.
This was a very exciting discovery, beyond the idea that this could be therapeutic strategy. It's very interesting that the ecDNA hub is really a completely different kind of a beast in terms of nuclear structure. We can think about a chromosome as a very long piece pf DNA with lots of proteins “decorating” on top of it, like histones and transcription factors. The ecDNA hub is actually the opposite: there's a protein core with individual DNA molecules decorating on top. So it can be thought of as the opposite organization!
PIW: Wow, that is really interesting to think about. And what might these discoveries about ecDNA hubs say about how cancers develop?
HYC: Because we observed all these indications of cooperative function between distinct ecDNAs, I think it says a lot about how ecDNAs undergo selection. An individual ecDNA doesn’t need to contain all the required regulatory elements and co-selection of ecDNAs may occur.
Because ecDNAs don't have centromeres, when a cancer cell divides there is random segregation; one daughter cell can get more copies of the ecDNA than the other. And if that ecDNA has a gene that makes the cancer cell grow better, then that cell will survive and it can very quickly jack up the copy number. If there's nothing useful on that ecDNA, you can very quickly lose it. So these ecDNAs are a rapid way for cancer cells to gain a fitness advantage.
PIW: Are these findings exclusive to glioblastoma and other specific cancer cell lines that were used in these studies? Can we expect these mechanisms to be happening in many other cancers?
RV: EcDNAs can be detected across a wide range of cancer types, including but not limited to breast cancer, lung cancer, and ovarian cancer. Given the consistency of our observations between our glioblastoma and prostate cancer models, we anticipate our findings of the functioning of ecDNA elements as mobile enhancers will translate to many other cancer types in which ecDNAs can be detected.
PIW: What do these oncogenic roles for ecDNAs mean for patients?
RV: Oncogenic ecDNAs are exclusively found in cancer cells and contribute to intratumoral heterogeneity and therapy resistance. Patients with ecDNA positive cancers have worse outcomes compared to those whose cancers do not contain ecDNA. We think ecDNA is demonstrating unique biological behavior, such as uneven segregation and functioning as mobile enhancers, which provide substantial rationale for the development of ecDNA-specific therapies.
HYC: Understanding the new biology of ecDNA brings hope for patients with cancers that are currently difficult to treat. As we learn more about unique dependencies of ecDNAs that are quite different from how genes operate on the normal 23 pairs of chromosomes, these dependencies are new targets for potential therapies to eradicate ecDNAs or stop the growth of ecDNA-containing tumors.
PIW: What are you particularly excited to work on next?
CLW: I’m excited to learn more about how ecDNAs replicate, find their chromosomal targets, and activate gene transcription. We are developing perturbation screens to do this.
RV: We are analyzing sequencing profiles of thousands of tumors to detect extrachromosomal DNA elements. In combination with single-cell based characterization of smaller cohorts, we’re continuing to identify the basic properties of ecDNAs in cancer.
HYC: We're really interested understanding more about intermolecular gene activation, as it is a new concept in mammalian gene regulation and is proving very important in cancer. Another aspect we’re really excited about is how ecDNAs are inherited when a cancer cell starts to divide and whether the ecDNA hubs contribute in some way. We’ve built these tools for live-cell imaging and we can actually make and watch movies of these events like mitosis!