RAS Pathway v3.0?
, by Frank McCormick
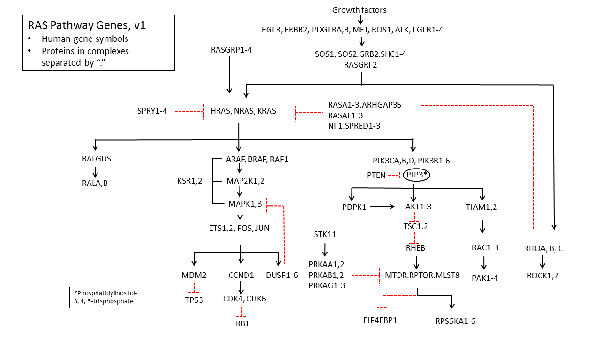
Two years ago I asked for your input on what genes comprise the "RAS Pathway". You responded with many suggestions that we incorporated into "v2.0" of the pathway, shown below. I'm happy to say that we relied heavily on your suggestions including collecting, synthesizing (when necessary), validating (according to TCGA) and sequencing all 180 genes in the v2 pathway. Through the community’s valuable input and the hard work of Dom Esposito and the Frederick National Laboratory for Cancer Research (FNLCR) RAS Reagents Group, the genes are now available through Addgene, in both the stop and no-stop versions. Considering that at least 20% of the ORF clones were either not available at all or were mis-annotated, this represents a significant contribution to our joint efforts to better understand how mutant RAS genes cause cancer.
Now, I think it's time we take another look at the pathway to determine if a version 3.0 is needed. What have we learned in the past two years about what genes and proteins modulate and feed into the RAS pathway? Are there any new genes to include for v3.0? Are there any in v2.0 that are less relevant now?
As always, we welcome your suggestions for changes in the pathway diagram. Provide your feedback by commenting below or email me at SolveRAS@nih.gov.