The RAS Initiative

More than 30 percent of all human cancers—including 95 percent of pancreatic cancers and 45 percent of colorectal cancers—are driven by mutations of the RAS family of genes. NCI established the RAS initiative in 2013 to explore innovative approaches for attacking the proteins encoded by mutant forms of RAS genes and to ultimately create effective, new therapies for RAS-related cancers.
-

RAS Research Teams
Learn about the nine highly collaborative research teams that compose the RAS Initiative. View their progress, projects, tools, collaborators, and team members.
-

RAS Tools and Resources
Find DNA reagents, cell line reagents, and protein production tools developed by researchers with the NCI RAS Initiative.
-
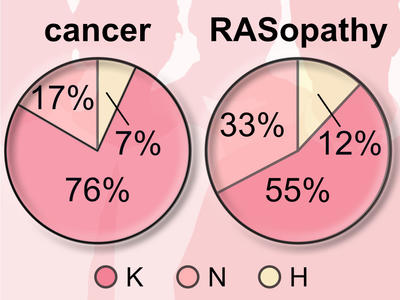
Publications from the RAS Initiative
This list of RAS Initiative publications reflects the balance between the public responsibilities of NCI and the requirements of external partners.
-
RAS News & Events
Through community and technical collaborations, workshops, and symposia, the RAS Initiative seeks to increase the sharing of knowledge and resources that are essential to defeat cancers caused by mutant RAS genes.
-
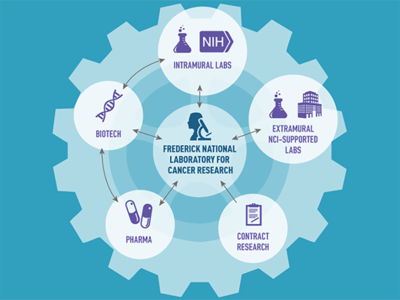
About the RAS Initiative
The "hub and spoke" model of the RAS Initiative connects researcher collaborators to better understand and target the more than 30% of cancers driven by mutations in RAS genes. Find more on the problem with RAS genes, RAS Initiative origins and oversight, and contact information.
-

RAS Dialogue Blog
Understanding and overcoming resistance to KRAS G12C inhibitors remains a key focus in the fight against RAS-driven cancers. In a new blog post, Drs. Mira and Nokin describe non-genetic methods of resistance to G12C inhibitors that bind to the inactive, GDP-bound form of the protein. They observed an increase in active GTP-loaded KRAS in patient samples, and argue for the dual inhibition of both active and inactive KRAS as a more effective therapeutic strategy.