Immunoprofiler Consortium at the University of California, San Francisco (UCSF)
, by Vincent Chan, Ph.D., Alexis Combes, Ph.D., Elizabeth Edmiston, Ph.D. and Matthew Krummel, Ph.D.
Cancer can be seen through a series of lenses. Historically, it was seen through the "tissue-of-origin lens" in which cancer was classified and treated based on where it had originated, such as in the skin, colon, or the kidneys. Subclassification of tissues-of-origin based on features was also prevalent, although not all were widely adopted. In breast cancer, for example, there are four major molecular subtypes: Luminal A, Luminal B, Triple-negative/Basal-like, and HER2-enriched. Therapies such as trastuzumab, a monoclonal antibody against HER2-enriched breast cancers, are aimed at these distinctive features. More recently, cancer was seen through the "oncogene lens," in which specific gene mutations are attributed to uncontrolled cell growth and proliferation observed in cancer. Targeted therapies were developed against oncogenes such as Ras, Myc, Rb, and BRAF. For example, vemurafenib is a small-molecule inhibitor of BRAF(V600E) kinase that demonstrates exceptional substrate specificity, and profound, yet often transient, effects on tumor size.
In the last few years, new cancer immunotherapy drugs have triggered remarkable and prolonged remissions in a subset of cancer patients. These therapies unleashed the patient's own immune system to locate and destroy cancer cells. The remarkable success of cancer immunotherapy as well as its limitations have underscored the need to view cancer through a new lens that takes immunology into account1. This has led to researchers, like us, to pursue cancer research through an "immunopathology lens". Through this lens, we've learned that there are also crucial differences among tumors and patients in the strength and durability of the immune response, and that we need a deeper understanding of this new dimension. Thus far, we have yet to fully understand even the most basic questions, including: How many distinct forms of immunopathologies exist in cancer? Do these immunopathologies cross tissue-of-origin and oncogene-driver boundaries? Can you have more than one immunopathology in a tumor? And what are the markers or signatures that identify them?
Through our and the research community's efforts, we are just beginning to reveal some of these distinct forms of immunopathologies in cancers 2,3. What we have outlined here are a few of the immunopathologies that may be present in combination within the tumor microenvironment, that we have yet to fully elucidate. “Immune privilege” is a situation in which immune response to pathogens is hindered to protect the organ function from damage by inflammatory immune reactions 4. A commonly observed “immune privilege” phenomenon in many types of cancers is exclusion of T cells from the vicinity of tumor nests so the tumor cells are protected 5. Many of these tumors are also poorly antigenic (see antigen) – unable to stimulate a strong T cell response against the tumor. Increasing evidence also suggests that the tumor microenvironment of many cancers potently suppress the immune response by activating multiple regulatory mechanisms such as the presence of inhibitory receptors, overabundance of regulatory T cells (Tregs), and imbalance of myeloid cells. “T cell exhaustion” is a broad term that can describe heterogeneous forms of distinct epigenetic and metabolic states of T cells, including reduced capacity to secrete cytokines and increased expression of inhibitory receptors. Recently, dysfunctional T cells that are chronically stimulated to partial or severe (irreversible) exhaustion were defined; first in chronic viral infections, and then in cancers.
The UCSF Immunoprofiler Consortium was launched with the goal of identifying distinct forms of immunopathologies associated with and across cancers. The Consortium was initially launched in January 2015 by UCSF and, shortly after, involved several large Biopharma companies as partners. The Consortium is presently a six-year initiative, surveying over 600 tumors and has matched adjacent normal tissues across more than 15 different forms of cancer.
The Consortium has also become a model for data sharing between academia and industry in what is now called “precompetitive data sharing”. The data and analysis over the course of this program aims to serve two major roles. First, by providing a better understanding of specific classes of immunopathology, it can guide the selection of patients and indications for existing and novel drugs. If a therapy treats a pathology, knowing which immunopathology is present guides the success of the therapy, especially an immunotherapy. Second, this study aims to discover targets for new therapeutic interventions. The cell types and gene expression that define specific types of tumor immune microenvironments (TIME) will serve to validate and direct new therapeutic efforts.
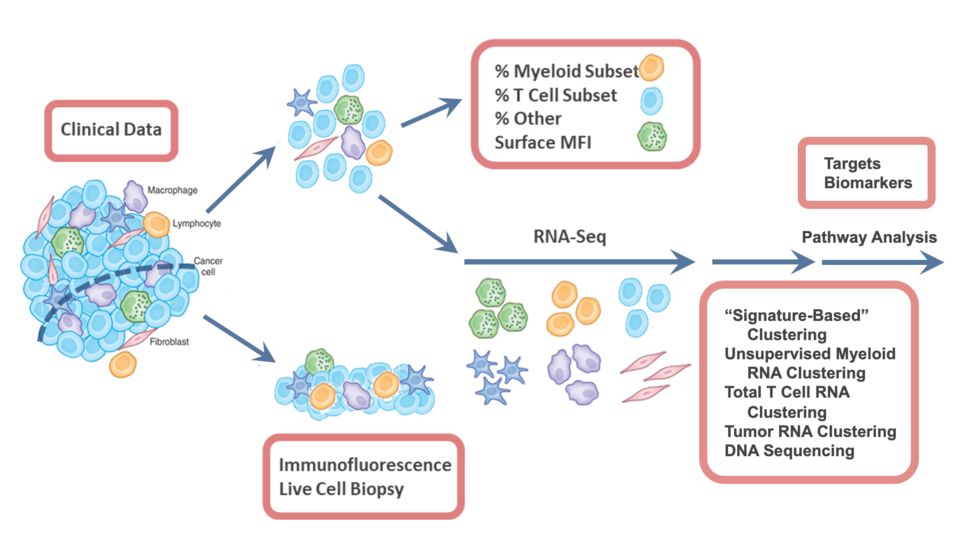
The overall scope of the Consortium is to coordinate the handling of valuable human biopsies and surgical resections taken from cancer patients, and to consistently perform multi-scale immunoprofiling assays — a series of tests that determine the tumor-immune composition, single-cell and population-level gene expression, and tumor-immune and immune-immune interaction biology. Shown in Figure 1 is the broad profiling trajectory that maximizes the use of each tumor and adjacent normal tissue. Once a tumor biopsy or surgical resection is removed from a patient, we immediately (<2 hours) transport it to the laboratory for processing. By taking it live and intact, we have the opportunity to study it much more intensely. A sliver or representative pie slice of the tumor is taken immediately for live tumor imaging to study tumor/immune cell interactions, or to be fixed for immunofluorescence imaging to study the spatial mapping of immune cells in tumors. The remaining tumor mass is then gently dissociated into single cells using an optimized standard protocol for digesting tumors to provide quantitative single cell analysis of the TIME. This method has been validated across multiple tumor types and tissues, and the numbers are in accordance with imaging.
The following standardized assays are performed across all tumors:
- Composition analysis based on multi-panel flow cytometry and/or CyTOF to measure the quantities and relative abundance of various cell types
- RNA sequencing of single cells and/or population-level (total live cells, T-cells, T-regs, myeloids, stromal cells, and tumor cells) to establish signatures of gene expression in tumors
- Whole exome sequencing to reveal the genetic mutations within tumors, with the aim of discovering genetic factors that may enhance or inhibit the immune response
Another essential component to the Immunoprofiler Consortium is a "human-to-mouse translator" program with the overall goal of providing a rational assignment of mouse models to their human counterparts. While mouse models have advanced our understanding of immune function and disease, they fail to account for the natural diversity in human immune responses. Therefore, we can imagine that a "translation table" between the immune profile of mouse models and their human counterparts will help researchers determine which mouse model to study, including two important environmental conditions: diet and age. First, the immune profiles of tumor models in mice will be benchmarked against data from the human tumor types. Second, mice will be assessed based on their high fat diet regimen and how it alters their antitumor immune response. Third, mice will be assessed based on their aged immune system and how it can impact the systemic and intratumoral immune composition in the host (and whether this brings specific models into greater similarity with specific classes of human disease).
The Consortium is unusual, but beneficial because of an explicit agreement among all members to share and have real-time access to the complete dataset. In turn, each of the Consortium members contributes a portion of the costs to collect the necessary biopsies and perform subsequent analyses. By sharing data at all levels, from biological discovery to drug development and patient prioritization, our ultimate goal is to accelerate the path to the next cures for all classes of cancer.
References
- Mujal AM, Krummel MF. Immunity as a continuum of archetypes. Science. 2019;364(6435):28-29. doi:10.1126/science. aau8694 (PMID: 30948539)
- Binnewies M, Mujal AM, Pollack JL, et al. Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4+ T Cell Immunity. Cell. 2019;177(3):556-571.e16. doi:10.1016/j.cell.2019.02.005 (PMID: 30955881)
- Barry KC, Hsu J, Broz ML, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24(8):1178-1191. doi:10.1038/s41591-018-0085-8 (PMID: 29942093)
- Hong S, Van Kaer L. Immune privilege: keeping an eye on natural killer T cells. J Exp Med. 1999;190(9):1197-1200. doi:10.1084/jem.190.9.1197 (PMID: 10544192)
- Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74-80. doi:10.1126/science.aaa6204 (PMID: 25838376)