Quantitative Assays for RAS Pathway Proteins and Phosphorylation States
, by Jim Hartley and Gordon Whiteley
In cooperation with the RAS Initiative, the NCI's Clinical Proteomic Tumor Analysis Consortium (CPTAC) has launched a project to develop quantitative assays for proteins and phosphopeptides involved in RAS signaling. Within the next 1-2 years these assays should allow the amounts and phosphorylation states of tens of RAS and RAS-related proteins to be determined in tumor samples, cell lines, or cancer models in a single run.
The initial target list for the CPTAC - RAS project is shown below. We invite your input on the best targets to improve our understanding of how the RAS pathway operates. Please submit a comment or send an email to SolveRAS@nih.gov.
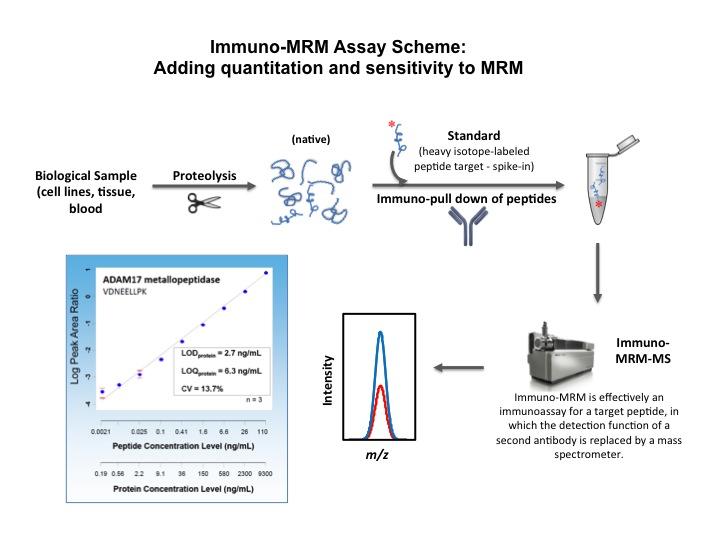
The capability to use mass spectrometry to determine the absolute amounts of peptides (and thus, proteins) is the culmination of years of developing concepts and technologies. First, pre-selected peptides derived from protein targets by trypsin digestion can be detected within a complex mixture using a targeted mass spectrometric technique called multiple reaction monitoring (MRM)1. Second, the quantities of these peptides in the mixture can be determined by spiking the samples with synthetic peptides of exactly the same amino acid sequences (and post-translational modifications, if appropriate) made distinguishable by the incorporation of heavy isotopes2. MRM assays can be developed to establish linear response curves, repeatability, lower limit of quantitation, etc. Third, in cases where protein targets are of low abundance, antibodies raised against their tryptic peptides can enrich for the target peptides and their spiked synthetic counterparts prior to MRM measurement, a process called immuno-MRM3. Fourth, both MRM and immuno-MRM assays can be highly multiplexed so that many peptide and protein targets can be determined from a single analytical run4. Finally, an extensive program of interlaboratory testing has established good reproducibility of these new techniques5.
The CPTAC program recently awarded contracts to the Fred Hutchinson Cancer Research Center (Dr. Amanda Paulovich), the Moffitt Cancer Center (Dr. John Koomen), and the Broad Institute (Dr. Steven Carr) to develop assays for the RAS pathway targets. The target list is being refined by comparing desired target peptides to databases of peptides that have already been detected in global proteomic screens.
The RAS pathway target list below comprises nearly 100 target peptides from 52 different proteins.
- Column A contains the names of the target proteins.
- Column B describes the kind of assay, either "Total" (measures the total amount of protein, using any convenient peptide), "P" (indicating that a specific phosphopeptide will be determined), or "non-P" (specifying a beta-catenin peptide important for regulating degradation).
- Column C gives the specific amino acid that is phosphorylated for the "P" targets in column B (for example, S473 for AKT1).
- Column D describes the class of each target, whether "upstream", "downstream", "effector", "regulator", or "RAS" itself. For some proteins several different assays for specific phosphopeptides will be developed (for example, assays for total BRAF protein and and BRAF phosphorylated at T401, S445, S465, and S467).The target list includes 11 RAS proteins: HRAS, NRAS, KRAS-4a, KRAS-4b, and six mutants of KRAS-4b.
| Target Protein | Specific Target | Phosporylation Site | Status |
|---|---|---|---|
| Akt1 | Total | Downstream | |
| Akt1 | P | S473 | Downstream |
| Akt1 | P | T308 | Downstream |
| Akt2 | Total | Downstream | |
| Akt3 | Total | Downstream | |
| Akt3 | P | S485 | Downstream |
| E-Cadherin | Total | Downstream | |
| N-Cadherin | Total | Downstream | |
| b-catenin | P | T41/S45 | Downstream |
| b-catenin | P | S552 | Downstream |
| b-catenin | non-P | (S33/S37/T41) | Downstream |
| PTEN | Total | Downstream | |
| PTEN | P | S380 | Downstream |
| ARAF | Total | Effector | |
| ARAF | P | S299 | Effector |
| BRAF | Total | Effector | |
| BRAF | P | T401 | Effector |
| BRAF | P | S467 | Effector |
| BRAF | P | S465 | Effector |
| BRAF | P | S445 | Effector |
| CRAF | Total | Effector | |
| CRAF | P | S259 | Effector |
| CRAF | P | S359 | Effector |
| CRAF | P | S338 | Effector |
| MEK1 | Total | Downstream | |
| MEK1 | P | S298 | Downstream |
| MEK1 | P | T386 | Downstream |
| MEK2 | Total | Downstream | |
| MEK2 | P | S221 | Downstream |
| mTOR | Total | Downstream | |
| mTOR | P | S2448 | Downstream |
| mTOR | P | S2481 | Downstream |
| GSK-b | Total | Downstream | |
| GSK-b | P | S9 | Downstream |
| ERK1/2 | Total | Downstream | |
| ERK1/2 | P | T202/Y204 | Downstream |
| c-Fos | Total | Downstream | |
| EGFR | Total | Upstream | |
| EGFR | P | S1046/1047 | Upstream |
| EGFR | P | Y1045 | Upstream |
| EGFR | P | Y1068 | Upstream |
| EGFR | P | Y1086 | Upstream |
| EGFR | P | Y1148 | Upstream |
| EGFR | P | Y1173 | Upstream |
| EGFR | P | Y845 | Upstream |
| EGFR | P | Y992 | Upstream |
| ErbB2 | Total | Upstream | |
| ErbB2 | P | Y1248 | Upstream |
| ErbB2 | P | Y877 | Upstream |
| ErbB3 | Total | Upstream | |
| ErbB3 | P | Y1289 | Upstream |
| ErbB4 | Total | Upstream | |
| ErbB4 | P | Y984 | Upstream |
| ErbB4 | P | Y1284 | Upstream |
| Cyclin D1 | Total | Downstream | |
| Cyclin D1 | P | T286 | Downstream |
| Raptor | Total | Downstream | |
| Raptor | P | S792 | Downstream |
| RasGRF1 | Total | Regulator | |
| RasGRF1 | P | S916 | Regulator |
| Rictor | Total | Downstream | |
| IRS-1 | Total | Upstream | |
| IRS-1 | P | S612 | Upstream |
| SPRED1 | Total | Regulator | |
| SPRED2 | Total | Regulator | |
| SPRED3 | Total | Regulator | |
| Sprouty1 | Total | Regulator | |
| Sprouty2 | Total | Regulator | |
| Sprouty3 | Total | Regulator | |
| Sprouty4 | Total | Regulator | |
| NF1 | Total | Regulator | |
| SYNGAP | Total | Regulator | |
| p120GAP | Total | Regulator | |
| RASGRP | Total | Regulator | |
| RASGRF2 | Total | Regulator | |
| Sos1 | Total | Regulator | |
| H-Ras | Total | RAS | |
| K-Ras 4A | Total | RAS | |
| K-Ras 4B | Total | RAS | |
| N-Ras | Total | RAS | |
| K-Ras 4B G12A | Total | RAS | |
| K-Ras 4B G12C | Total | RAS | |
| K-Ras 4B G12D | Total | RAS | |
| K-Ras 4B G12V | Total | RAS | |
| K-Ras 4B Q61L | Total | RAS | |
| K-Ras 4B Q61R | Total | RAS | |
| K-Ras 4B | P | S181 | RAS |
| Calmodulin | Total | Effector | |
| CaM kinase | Total | Downstream | |
| CaM kinase | P | S286 | Downstream |
| KSR1 | Total | Downstream | |
| KSR2 | Total | Downstream | |
| IQGAP | Total | Regulator | |
| DUSP1 | Total | Downstream | |
| DUSP2 | Total | Downstream | |
| DUSP4 | Total | Downstream | |
| DUSP6 | Total | Downstream | |
| DUSP7 | Total | Downstream |
Selected References
- Boja, ES, Rodriguez, H. Mass spectrometry-based targeted quantitative proteomics: achieving sensitive and reproducible detection of proteins. Proteomics. 2012; 12(8): 1093-110.
- Hoofnagle, AN, et al. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012; 58(4): 777-781.
- Kuhn, E, et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012; 11(6): M111.013854.
- Whiteaker JR, et al. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol Cell Proteomics. 2012; 11(6): M111.015347.
- Kennedy, JJ, et al., Nat Methods 11, 149, 2014, doi: 10.1038/nmeth.2763.