The RAS Reference Reagents Program: Sharing Resources and Enabling RAS Research
, by Dom Esposito
The RAS Reference Reagents (RRR) program within the RAS Initiative at the Frederick National Laboratory for Cancer Research has a twofold mission: first, to develop nucleic acid and protein reagents to support the needs of the various project teams within the RAS Initiative, and second to promote RAS-related research in the external community. Here we describe this second mission.
Independent research efforts toward a common objective are essential for robust scientific results. However, two weaknesses are inherent in this organizational approach. First, it may be difficult to compare results generated with cell lines, DNA constructs, proteins, or other reagents constructed or maintained in different labs. Contradictory results from this discordance cause confusion in the community and slow progress. Second, multiple labs may spend time and money constructing the same reagents over and over again. For example, a quick scan of online DNA repositories such as Addgene will demonstrate that for many genes (p53 for instance) there are dozens of similar or identical constructs generated by multiple laboratories. A goal of the RAS Initiative is to generate a set of validated reference reagents and protocols to minimize both of these issues.
The first step for the RRR Program was to generate DNA reagents to support various internal RAS Initiative projects. These reagents are now available to the external community in our initial clone collections. These include a series of KRAS clones with various oncogenic mutations, and a series of RAS pathway genes of interest including GAPs, GEFs, and effectors. In order to gain the most utility from these clones, our laboratory uses the Gateway recombination-based cloning system in conjunction with a combinatorial cloning platform to permit generation of a large number of DNA reagents from a small set of Entry clones. The RRR clone collections contain genes with and without stop codons so that fusion constructs can be made at either end of the proteins of interest. This arrangement enables a wide variety of tagging schemes including solubility tags for protein production, fluorescent tags for localization, and epitope tags for detection or immunoprecipitation.
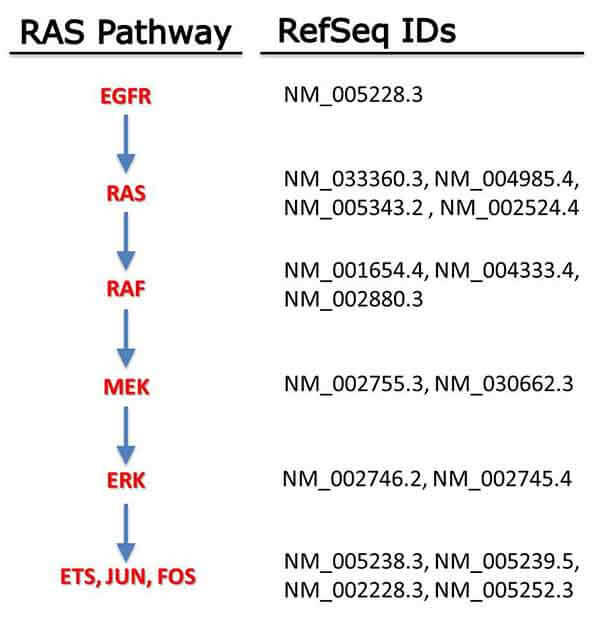
In the process of generating these initial clone sets, we decided it would also be worthwhile to examine a larger set of RAS-associated genes. Fortunately, around this time a discussion was started on this website to identify a consensus set of RAS pathway genes. This list has now grown to nearly 200 genes, and we began to assess these for development of a larger set of clones. Interestingly, a number of bioinformatic challenges arose during this process which have implications for RAS research. While we expected to deal with splice variants, it was surprising that good bioinformatic data on what comprised the “best” or “most common” transcript variants of many widely-studied RAS-related proteins were lacking. For example, the sequences in NCBI’s RefSeq library are predominantly the longest identified transcript of each gene, but comparisons to expression data from TCGA and other sources show that the longest transcripts are often not the ones that are responsible for important phenotypes. In some cases, a significant literature exists for transcript variants that are dramatically underexpressed in human tissues, suggesting that these are probably not the best model systems for studying those genes. In other cases, the most expressed transcript of a given gene is not found in commercial cDNA libraries at all, forcing researchers either to work on the available splice variants with questionable biological relevance, or to dedicate resources and time to make the correct clones. Overall, of the 181 genes that we surveyed, more than 20% lack commercially available cDNA or ORFeome clones for the transcript variant which is most commonly expressed. It is our goal as part of the next step of the RRR Program to acquire or generate ORF clones of all of the 181 genes in the RAS pathway in their most biologically relevant form. For the reasons described above, such a library should be a significant benefit to the RAS community.
Ultimately, some of the more challenging reagents to generate are purified, highly qualified proteins. Even after the properties of each protein are well understood, there is usually a significant cost to producing them, and the level of expertise required is not trivial. Thus, the RAS Initiative at FNLCR does not have as a goal the generation of large quantities of proteins for distribution, but we do want to provide the raw materials and validated protocols to allow researchers to generate their own RAS and RAS-related proteins. Our hope is that the community can take advantage of the tricks that we’ve learned and the pitfalls we’ve encountered to make the process of generating protein reagents easier and faster. To this end, we hope soon to be able to provide DNA reagents, cell lines, and viruses for production of various RAS-related proteins, along with detailed protocols on the purification and QC of these reagents.
We would greatly appreciate any suggestions or feedback on our RRR program.
About the Author
Dom Esposito has a background in determining structure-function relationships of recombination proteins. During a stint in biotech he helped develop Gateway cloning. His responsibilities in the NCI RAS Initiative include management of the RAS Reference Reagents Group.